rsEGFP2 WT Switching QY#
ON->OFF#
Load Experimental Switching Data#
We begin by importing the required library and loading the actinometry measurement data from a CSV file. The dataset is stored in exp_data for further analysis
import pandas as pd
exp_data = pd.read_csv("DATA/prot1 ON2OFF.csv")
exp_data
| timestamp | cycle | type | 186.85486 | 187.31995223015844 | 187.78500323297956 | 188.250012996982 | 188.71498151068454 | 189.17990876260572 | 189.64479474126426 | ... | 1032.9632529736894 | 1033.3204920667242 | 1033.6776665334785 | 1034.034776362471 | 1034.3918215422202 | 1034.7488020612445 | 1035.1057179080635 | 1035.4625690711953 | 1035.8193555391586 | 1036.176077300472 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 2025-04-04 13:05:43.323760 | 1 | zero | 16.990556 | 24978.543889 | 59.831028 | 47.330833 | 62.015528 | 71.481694 | 76.942944 | ... | 470.274306 | 455.953694 | 458.259556 | 465.662583 | 459.958611 | 461.657667 | 467.847083 | 466.512111 | 466.512111 | 466.512111 |
| 1 | 2025-04-04 13:06:24.439856 | 1 | on | 16.990556 | 24978.543889 | 60.195111 | 42.233667 | 55.097944 | 67.234056 | 57.525167 | ... | 457.774111 | 464.084889 | 463.114000 | 457.288667 | 441.511722 | 455.346889 | 469.182056 | 456.560500 | 456.560500 | 456.560500 |
| 2 | 2025-04-04 13:06:25.472725 | 1 | on | 16.990556 | 24978.543889 | 57.174568 | 48.544444 | 41.802160 | 43.150617 | 67.692531 | ... | 453.081481 | 459.554074 | 460.632840 | 454.429938 | 472.229568 | 473.038642 | 461.981296 | 435.821235 | 435.821235 | 435.821235 |
| 3 | 2025-04-04 13:06:26.506516 | 1 | on | 16.990556 | 24978.543889 | 74.030278 | 60.437833 | 58.981500 | 54.369778 | 53.641611 | ... | 462.871278 | 474.036500 | 470.881111 | 459.230444 | 455.346889 | 477.434611 | 456.075056 | 451.463333 | 451.463333 | 451.463333 |
| 4 | 2025-04-04 13:06:27.537642 | 1 | on | 16.990556 | 24978.543889 | 55.826111 | 47.195988 | 54.207963 | 80.907407 | 69.850062 | ... | 471.690185 | 486.523210 | 459.823765 | 473.847716 | 457.126852 | 462.520679 | 440.945370 | 446.878580 | 446.878580 | 446.878580 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 386 | 2025-04-04 13:13:02.957585 | 1 | on | 16.990556 | 24978.543889 | 34.250802 | 37.756790 | 59.601790 | 63.916852 | 55.826111 | ... | 435.281852 | 464.138827 | 449.575494 | 446.878580 | 447.957346 | 459.554074 | 450.384568 | 449.845185 | 449.845185 | 449.845185 |
| 387 | 2025-04-04 13:13:03.987853 | 1 | on | 16.990556 | 24978.543889 | 58.792716 | 15.372407 | 35.599259 | 54.747346 | 83.604321 | ... | 463.599444 | 464.138827 | 464.138827 | 454.969321 | 460.363148 | 433.124321 | 450.923951 | 447.417963 | 447.417963 | 447.417963 |
| 388 | 2025-04-04 13:13:05.020339 | 1 | on | 16.990556 | 24978.543889 | 30.475123 | 42.880926 | 57.983642 | 69.040988 | 55.017037 | ... | 471.150802 | 478.432469 | 456.048086 | 444.990741 | 465.217593 | 461.172222 | 452.002716 | 445.260432 | 445.260432 | 445.260432 |
| 389 | 2025-04-04 13:13:06.054197 | 1 | on | 16.990556 | 24978.543889 | 53.129198 | 22.923765 | 36.678025 | 38.835556 | 69.580370 | ... | 438.248457 | 441.754444 | 446.339198 | 457.396543 | 455.239012 | 449.305802 | 435.281852 | 457.935926 | 457.935926 | 457.935926 |
| 390 | 2025-04-04 13:13:19.090390 | 1 | static | 16.990556 | 24978.543889 | 44.175444 | 36.772417 | 47.330833 | 41.505500 | 62.015528 | ... | 439.812667 | 440.176750 | 464.813056 | 451.584694 | 446.851611 | 466.269389 | 448.672028 | 451.706056 | 451.706056 | 451.706056 |
391 rows × 2051 columns
Preprocess Spectral Data and Compute Absorbance#
We now prepare the spectral intensity data for analysis. The dataset contains different types of measurements:
“zero”: baseline measurements,
“static”: background noise or dark reference,
Other types: actual actinometry measurements over time.
We extract these components, convert timestamps, and compute the absorbance spectrum using the formula:
$$A(\lambda) = -\log_{10} \left( \frac{I(\lambda) - I_{\text{static}}(\lambda)}{I_{\text{zero}}(\lambda) - I_{\text{static}}(\lambda)} \right)$$
This normalization corrects for both dark current and reference intensity variations.
import numpy as np
intensities=np.array(exp_data[(exp_data["type"] != "zero") & (exp_data["type"] != "static")].iloc[:, 3:], dtype=np.float64)
static=np.array(exp_data[(exp_data["type"]== "static")].iloc[:, 3:], dtype=np.float64)[0]
zero=np.array(exp_data[(exp_data["type"]== "zero")].iloc[:, 3:], dtype=np.float64)[0]
wavelengths = np.array(exp_data.columns[3:], dtype=np.float64)
timestamps = pd.to_datetime(exp_data["timestamp"][(exp_data["type"] != "zero") & (exp_data["type"] != "static")]) # Convert timestamp strings to datetime objects
timestamps = np.array((timestamps - timestamps.iloc[0]).dt.total_seconds()) # Convert to seconds since the first timestamp
def compute_absorbance(intensities: np.ndarray, static: np.ndarray, zero: np.ndarray) -> np.ndarray:
EPS = 1e-12
num = intensities - static
den = np.maximum(zero - static, EPS) # Éviter division par zéro
absorbance = -np.log10(np.maximum(num / den, EPS)) # Éviter log(0) ou log(négatif)
return absorbance
absorbance = compute_absorbance(intensities, static, zero)
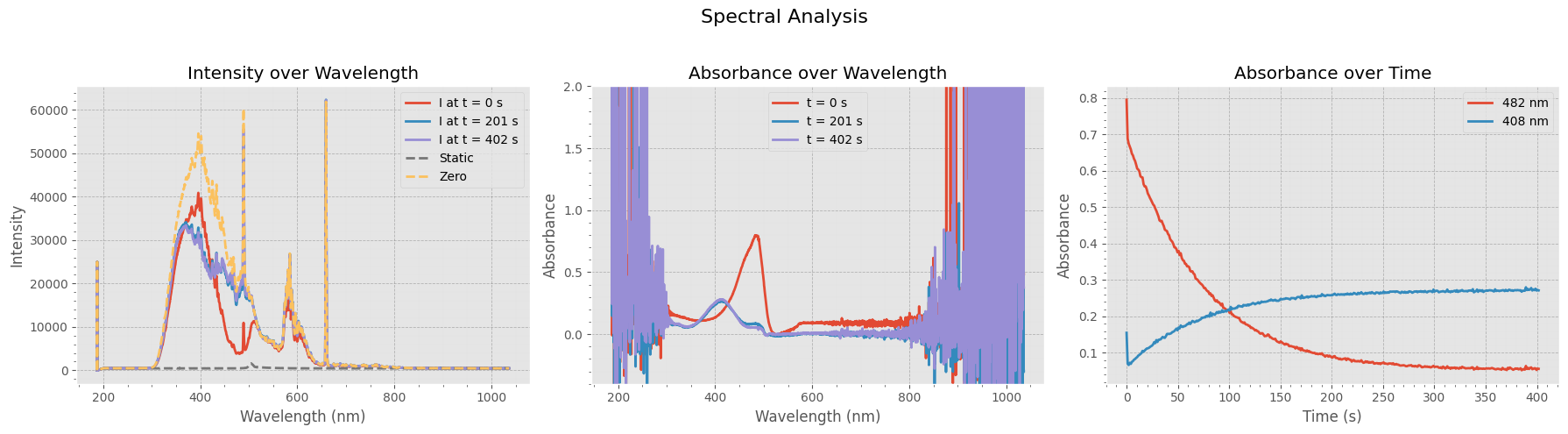
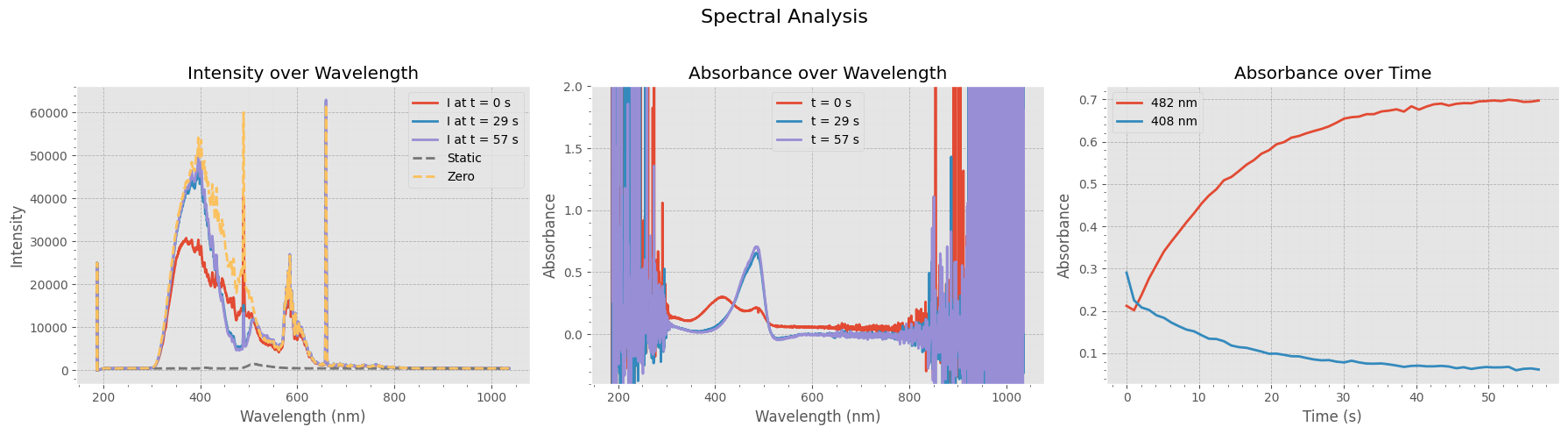
Visualizing Spectral Intensity and Absorbance#
This figure provides a comprehensive view of the switching data:
Left Panel: Raw intensity spectra at three time points, along with the static and zero references.
Middle Panel: Corresponding absorbance spectra at the same time points, showing how absorption evolves over time.
Right Panel: Absorbance at two key wavelengths (482 nm and 408 nm) plotted as a function of time, highlighting dynamic changes in the sample.
These plots help assess the stability and dynamics of the measured signals and the quality of the zero/static references.
Show code cell source
import matplotlib.pyplot as plt
import matplotlib.cm as cm
# Set global style
plt.style.use("ggplot") # Clean style
fig, axs = plt.subplots(1, 3, figsize=(18, 5))
fig.suptitle("Spectral Analysis", fontsize=16)
# --- Intensity Plot ---
axs[0].plot(wavelengths, intensities[0, :], label=f"I at t = {timestamps[0]:.0f} s", linewidth=2)
axs[0].plot(wavelengths, intensities[len(intensities)//2, :], label=f"I at t = {timestamps[len(intensities)//2]:.0f} s", linewidth=2)
axs[0].plot(wavelengths, intensities[-1, :], label=f"I at t = {timestamps[-1]:.0f} s", linewidth=2)
axs[0].plot(wavelengths, static, '--', label="Static", linewidth=2)
axs[0].plot(wavelengths, zero, '--', label="Zero", linewidth=2)
axs[0].set_title("Intensity over Wavelength")
axs[0].set_xlabel("Wavelength (nm)")
axs[0].set_ylabel("Intensity")
axs[0].legend()
axs[0].grid(True, which='major', linestyle='--', linewidth=0.6, color='gray', alpha=0.5, zorder=0)
axs[0].minorticks_on()
axs[0].grid(True, which='minor', linestyle=':', linewidth=0.3, color='lightgray', alpha=0.4, zorder=0)
# --- Absorbance Plot ---
axs[1].plot(wavelengths, absorbance[0, :], label=f"t = {timestamps[0]:.0f} s", linewidth=2)
axs[1].plot(wavelengths, absorbance[len(absorbance)//2, :], label=f"t = {timestamps[len(absorbance)//2]:.0f} s", linewidth=2)
axs[1].plot(wavelengths, absorbance[-1, :], label=f"t = {timestamps[-1]:.0f} s", linewidth=2)
axs[1].set_title("Absorbance over Wavelength")
axs[1].set_xlabel("Wavelength (nm)")
axs[1].set_ylabel("Absorbance")
axs[1].set_ylim(-0.4, 2)
axs[1].legend()
axs[1].grid(True, which='major', linestyle='--', linewidth=0.6, color='gray', alpha=0.5, zorder=0)
axs[1].minorticks_on()
axs[1].grid(True, which='minor', linestyle=':', linewidth=0.3, color='lightgray', alpha=0.4, zorder=0)
# --- Absorbance vs Time Plot ---
WL = [482, 408]
idxs = [np.argmin(np.abs(wavelengths - wl)) for wl in WL]
for i, idx in enumerate(idxs):
axs[2].plot(timestamps, absorbance[:, idx], label=f"{wavelengths[idx]:.0f} nm", linewidth=2)
axs[2].set_title("Absorbance over Time")
axs[2].set_xlabel("Time (s)")
axs[2].set_ylabel("Absorbance")
axs[2].legend()
axs[2].grid(True, which='major', linestyle='--', linewidth=0.6, color='gray', alpha=0.5, zorder=0)
axs[2].minorticks_on()
axs[2].grid(True, which='minor', linestyle=':', linewidth=0.3, color='lightgray', alpha=0.4, zorder=0)
plt.tight_layout(rect=[0, 0, 1, 0.95])
plt.show()
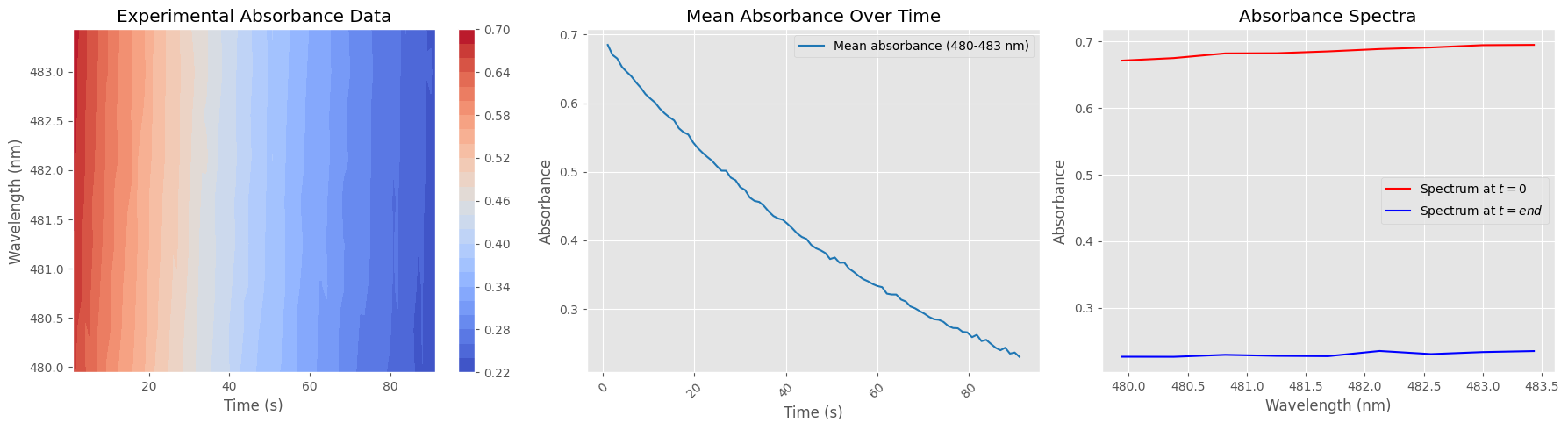
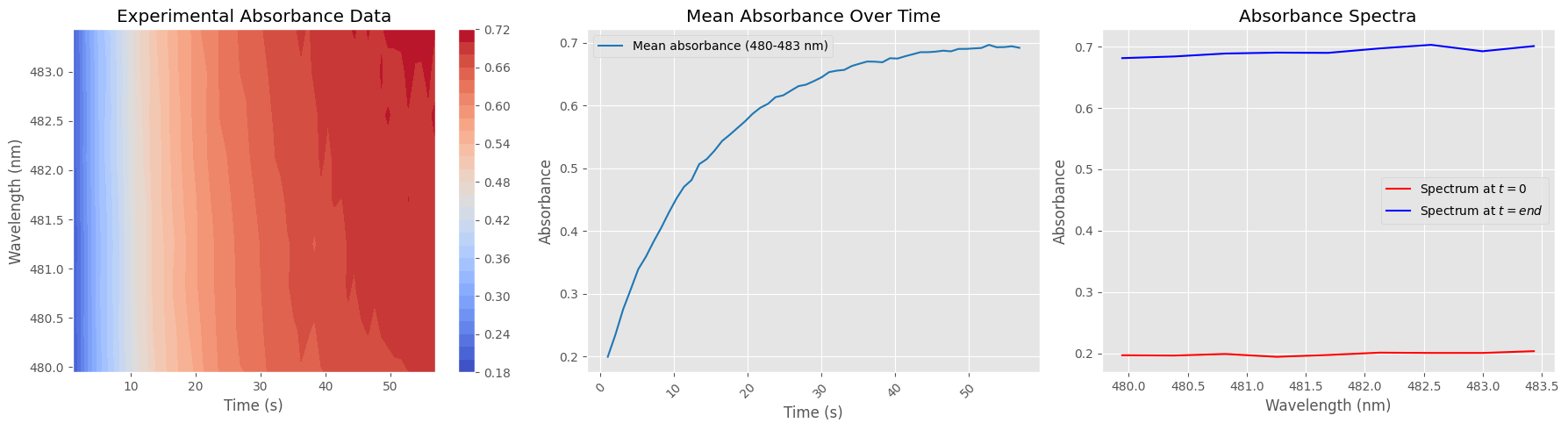
Data Cleaning and Absorbance Subset Preparation#
This cell prepares the dataset for focused analysis by:
Selecting a narrow wavelength range (e.g. 480–484 nm)
Normalizing absorbance values
Removing unwanted time points (first spectrum and last 300)
The resulting cleaned subset is suitable for targeted visualization or kinetic analysis.
# Define wavelength range of interest
wavelength_range = [480, 484]
idx_range = np.argmin(np.abs(wavelengths[:, None] - wavelength_range), axis=0)
# Extract and normalize absorbance data within the wavelength range
absorbance_subset = absorbance[:, idx_range[0]:idx_range[1]]
# absorbance_subset -= np.min(absorbance_subset)
wavelengths_subset = wavelengths[idx_range[0]:idx_range[1]]
# Filter out specific time points (e.g., first frame and last 300 entries)
# Adjust indices as needed
time_filter_indices = [0] + list(range(-300, 0))
absorbance_subset = np.delete(absorbance_subset, time_filter_indices, axis=0)
timestamps_filtered = np.delete(timestamps, time_filter_indices)
# Create subplots
fig, axs = plt.subplots(1, 3, figsize=(18, 5))
# --- 1. Absorbance Heatmap ---
X, Y = np.meshgrid(timestamps_filtered, wavelengths_subset)
contour = axs[0].contourf(X, Y, absorbance_subset.T, levels=30, cmap="coolwarm")
axs[0].set_title("Experimental Absorbance Data")
axs[0].set_xlabel("Time (s)")
axs[0].set_ylabel("Wavelength (nm)")
fig.colorbar(contour, ax=axs[0])
# --- 2. Absorbance Over Time (Averaged Across Wavelength Range) ---
mean_absorbance = np.mean(absorbance_subset, axis=1)
axs[1].plot(timestamps_filtered, mean_absorbance, label=f"Mean absorbance ({wavelengths_subset[0]:.0f}-{wavelengths_subset[-1]:.0f} nm)", color='tab:blue')
axs[1].set_title("Mean Absorbance Over Time")
axs[1].set_xlabel("Time (s)")
axs[1].set_ylabel("Absorbance")
axs[1].tick_params(axis="x", rotation=45)
axs[1].legend()
axs[1].grid(True)
# --- 3. Absorbance Spectrum at Start and End ---
axs[2].plot(wavelengths_subset, absorbance_subset[0, :], label="Spectrum at $t=0$", color='red')
axs[2].plot(wavelengths_subset, absorbance_subset[-1, :], label="Spectrum at $t=end$", color='blue')
axs[2].set_title("Absorbance Spectra")
axs[2].set_xlabel("Wavelength (nm)")
axs[2].set_ylabel("Absorbance")
axs[2].legend()
axs[2].grid(True)
plt.tight_layout()
plt.show()
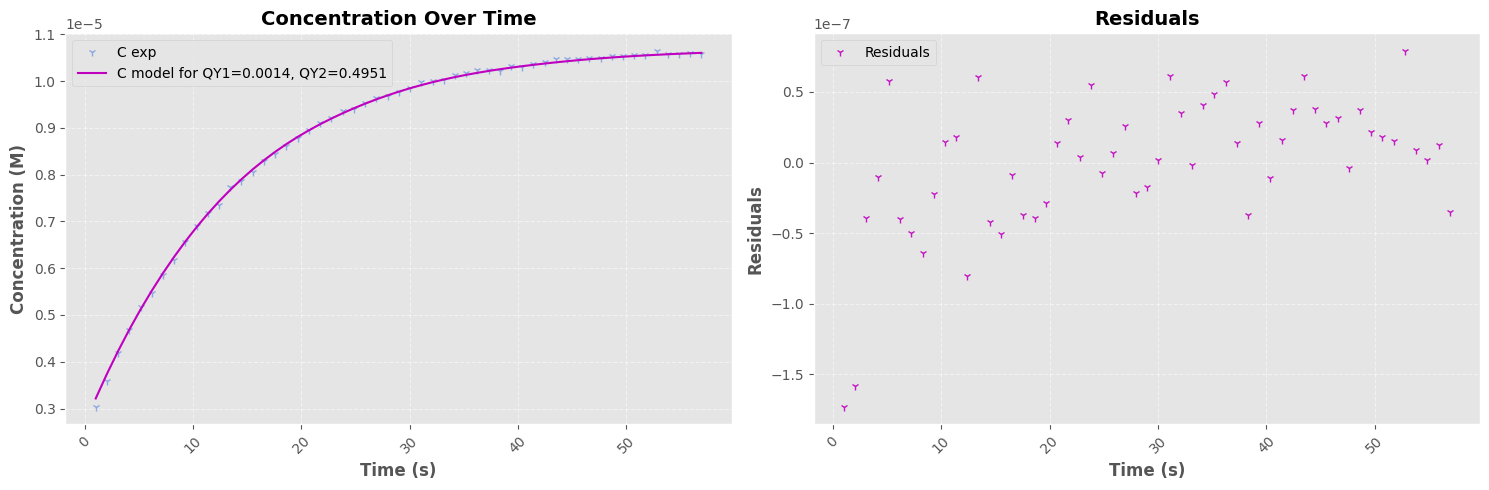
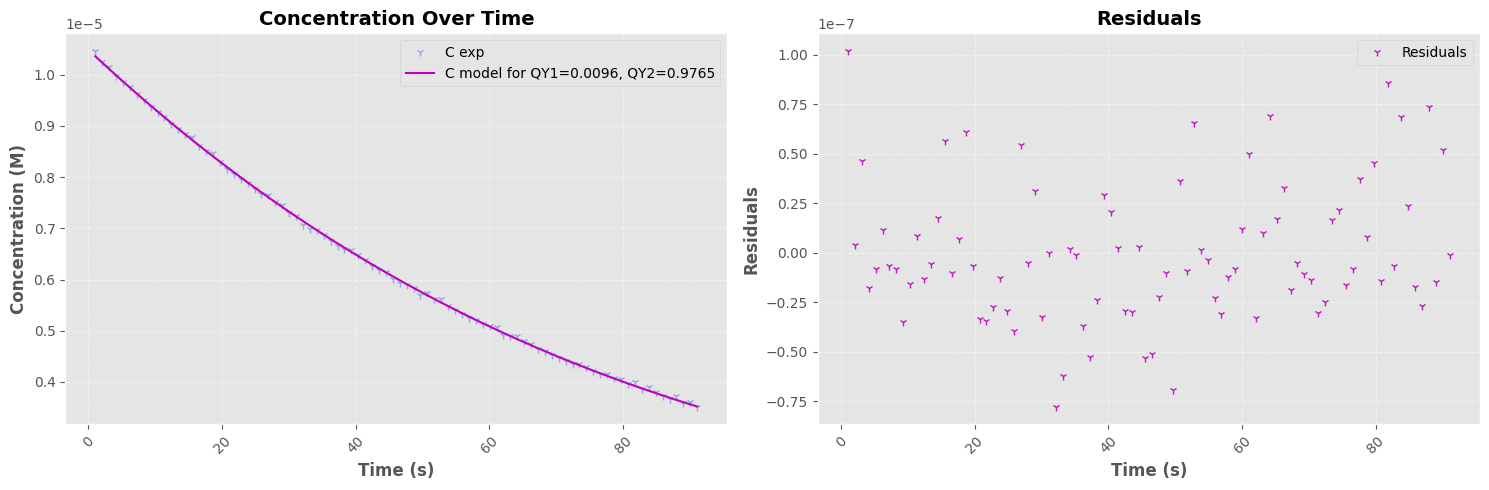
QY ON->OFF Regression#
from scipy.integrate import solve_ivp
from scipy.optimize import curve_fit
import numpy as np
import matplotlib.pyplot as plt
# === Physics constants ===
h = 6.62607004 * 10 ** (-34) # Planck's constant (J·s)
NA = 6.02214086 * 10 ** (23) # Avogadro's number (mol^-1)
c_vaccum = 299792458 # Speed of light in vacuum (m/s)
wl = 505 # Wavelength (nm)
v = c_vaccum / (wl * 1e-9) # Frequency (Hz)
volume = 23e-4 # Volume (L)
l = 1 # Path length (cm)
eps_on_482 = 65474 # Molar absorptivity (L·mol^-1·cm^-1)
eps_off_482 = 65 # Molar absorptivity (L·mol^-1·cm^-1)
eps_on_505 = 0.4166666667 * eps_on_482 # Adjusted molar absorptivity at 505
eps_off_505 = 0.4166666667 * eps_off_482 # Adjusted molar absorptivity at 505
I_w_list = [14.27e-3, 10.4e-3] # Irradiance values (W/m^2)
# === Experimental data ===
# Assuming 'timestamps_filtered' and 'C_exp' are already defined
# === Initialize parameters ===
lpopt = []
for I_w in I_w_list:
# Compute initial concentration based on experimental data
C_exp = mean_absorbance / (eps_on_482 * l)
C0 = C_exp[0]
I_0 = I_w / (h * v * NA) / volume # Photon irradiance (mol·L^-1·s^-1)
# === ODE definition ===
def dC_dt(t, C_ON, QY_ON2OFF, QY_OFF2ON):
C_OFF = C0 - C_ON
Iabs_ON = I_0 * (1 - np.exp(-eps_on_505 * C_ON * l * np.log(10)))
Iabs_OFF = I_0 * (1 - np.exp(-eps_off_505 * C_OFF * l * np.log(10)))
dCCF_deri = -QY_ON2OFF * Iabs_ON + QY_OFF2ON * Iabs_OFF
return dCCF_deri
# === Fitting function wrapper ===
def model(t, QY_ON2OFF, QY_OFF2ON, offset):
sol = solve_ivp(dC_dt, [t[0], t[-1]], [C0], args=(QY_ON2OFF, QY_OFF2ON), t_eval=t)
if not sol.success:
print("⚠️ ODE solver failed:", sol.message)
return np.full_like(t, np.nan)
return sol.y[0]+ offset
# === Fit the model to the experimental data ===
initial_guess = (0.001, 0.01, 0) # QY_ON2OFF, QY_OFF2ON, offset
bounds = ([1e-4, 1e-4,-1], [0.2, 1,1]) # ([min1, min2, ...], [max1, max2, ...])
popt, pcov = curve_fit(model, timestamps_filtered, C_exp, p0=initial_guess, bounds=bounds)
# Extract fitted parameters and compute uncertainties
uncertainties = np.sqrt(np.diag(pcov))
lpopt.append(popt)
print(f"=== Fit Results I={I_w:0.3e} W ===")
for i, (coef, incert) in enumerate(zip(popt, uncertainties), start=1):
print(f"Parameter {i}: {coef:.4f} ± {incert:.4f}")
# Compute fitted values
fitted_values = model(timestamps_filtered, *popt)
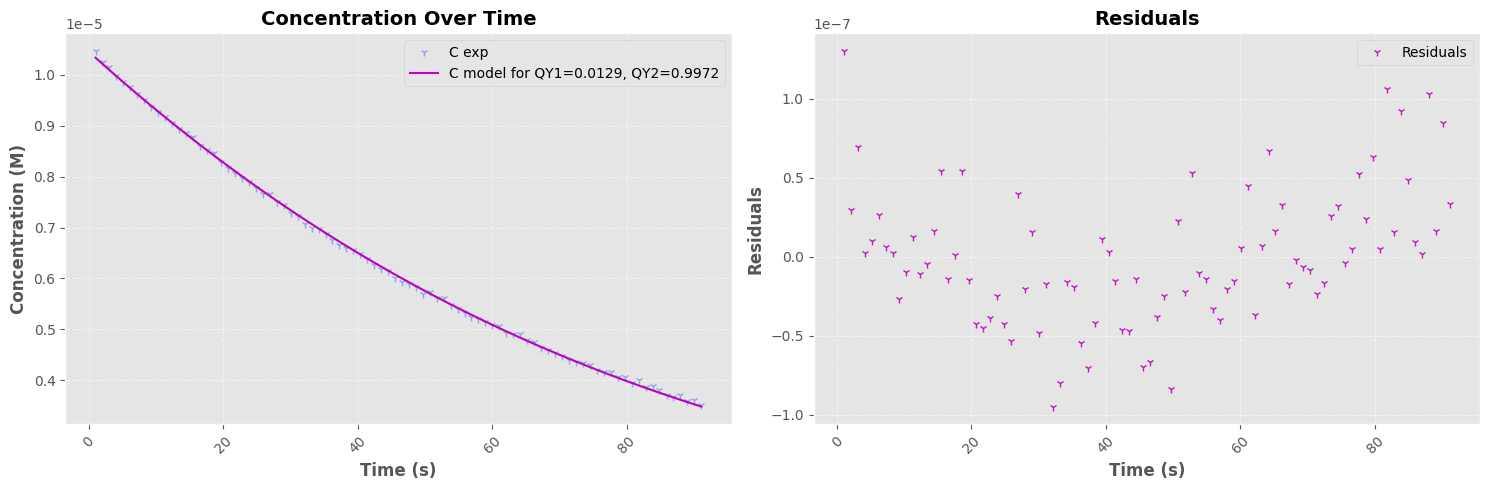
# === Plotting ===
fig, a = plt.subplots(1, 2, figsize=(15, 5))
pastel_colors = ["m", "#f9955e", "#5080e0"] # Mauve, orange, sky blue
# First Plot - Concentration Over Time
a[0].plot(timestamps_filtered, C_exp, "1", color=pastel_colors[2], label="C exp", alpha=0.6)
a[0].plot(timestamps_filtered, fitted_values, "-", color=pastel_colors[0], label=f"C model for QY1={popt[0]:0.4f}, QY2={popt[1]:0.4f}")
a[0].set_xlabel("Time (s)", fontsize=12, fontweight="bold")
a[0].set_ylabel("Concentration (M)", fontsize=12, fontweight="bold")
a[0].set_title("Concentration Over Time", fontsize=14, fontweight="bold")
a[0].tick_params(axis="x", rotation=45)
a[0].grid(linestyle="--", alpha=0.5)
a[0].legend()
# Second Plot - Residuals Over Time
residuals = C_exp - fitted_values
a[1].plot(timestamps_filtered, residuals, "1", color=pastel_colors[0], label="Residuals", alpha=0.9)
a[1].set_xlabel("Time (s)", fontsize=12, fontweight="bold")
a[1].set_ylabel("Residuals", fontsize=12, fontweight="bold")
a[1].set_title("Residuals", fontsize=14, fontweight="bold")
a[1].tick_params(axis="x", rotation=45)
a[1].grid(linestyle="--", alpha=0.5)
a[1].legend()
plt.tight_layout()
plt.show()
=== Fit Results I=1.427e-02 W ===
Parameter 1: 0.0096 ± 0.0001
Parameter 2: 0.9765 ± 0.0604
Parameter 3: -0.0000 ± 0.0000
=== Fit Results I=1.040e-02 W ===
Parameter 1: 0.0129 ± 0.0001
Parameter 2: 0.9972 ± 0.1030
Parameter 3: -0.0000 ± 0.0000
OFF -> ON#
Load Experimental Switching Data#
We begin by importing the required library and loading the actinometry measurement data from a CSV file. The dataset is stored in exp_data for further analysis
import pandas as pd
exp_data = pd.read_csv("DATA/prot1 OFF2ON.csv")
exp_data
| timestamp | cycle | type | 186.85486 | 187.31995223015844 | 187.78500323297956 | 188.250012996982 | 188.71498151068454 | 189.17990876260572 | 189.64479474126426 | ... | 1032.9632529736894 | 1033.3204920667242 | 1033.6776665334785 | 1034.034776362471 | 1034.3918215422202 | 1034.7488020612445 | 1035.1057179080635 | 1035.4625690711953 | 1035.8193555391586 | 1036.176077300472 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 2025-04-04 13:18:08.282867 | 1 | zero | 16.990556 | 24978.543889 | 33.859750 | 28.155778 | 33.738389 | 40.898694 | 38.956917 | ... | 449.157472 | 435.807750 | 441.026278 | 435.443667 | 437.021361 | 448.672028 | 442.968056 | 451.463333 | 451.463333 | 451.463333 |
| 1 | 2025-04-04 13:18:36.280977 | 1 | off | 16.990556 | 24978.543889 | 45.038457 | 22.114691 | 31.823580 | 32.093272 | 49.623210 | ... | 441.215062 | 443.642284 | 457.935926 | 446.339198 | 431.506173 | 445.530123 | 440.405988 | 450.384568 | 450.384568 | 450.384568 |
| 2 | 2025-04-04 13:18:37.311192 | 1 | off | 16.990556 | 24978.543889 | 31.823580 | 22.114691 | 42.341543 | 53.938272 | 58.792716 | ... | 450.114877 | 450.384568 | 433.933395 | 450.923951 | 459.823765 | 444.721049 | 440.136296 | 459.823765 | 459.823765 | 459.823765 |
| 3 | 2025-04-04 13:18:38.352852 | 1 | off | 16.990556 | 24978.543889 | 54.477654 | 32.093272 | 37.487099 | 50.162593 | 59.332099 | ... | 440.675679 | 443.372593 | 440.405988 | 445.530123 | 430.966790 | 437.439383 | 452.272407 | 429.078951 | 429.078951 | 429.078951 |
| 4 | 2025-04-04 13:18:39.383552 | 1 | off | 16.990556 | 24978.543889 | 39.644630 | 36.947716 | 37.756790 | 31.284198 | 61.219938 | ... | 425.033580 | 463.599444 | 445.530123 | 451.193642 | 443.642284 | 453.620864 | 435.012160 | 434.203086 | 434.203086 | 434.203086 |
| 5 | 2025-04-04 13:18:40.416396 | 1 | off | 16.990556 | 24978.543889 | 19.687469 | 33.172037 | 42.341543 | 18.608704 | 38.296173 | ... | 453.620864 | 446.878580 | 429.888025 | 438.518148 | 424.494198 | 439.327222 | 434.203086 | 451.193642 | 451.193642 | 451.193642 |
| 6 | 2025-04-04 13:18:41.449506 | 1 | off | 16.990556 | 24978.543889 | 28.317593 | 28.317593 | 30.205432 | 32.093272 | 42.611235 | ... | 461.172222 | 433.933395 | 444.181667 | 432.315247 | 436.900000 | 419.909444 | 459.014691 | 448.766420 | 448.766420 | 448.766420 |
| 7 | 2025-04-04 13:18:42.486227 | 1 | off | 16.990556 | 24978.543889 | 40.453704 | 46.656605 | 20.766235 | 49.623210 | 65.265309 | ... | 446.878580 | 450.114877 | 426.382037 | 429.348642 | 425.842654 | 437.169691 | 444.990741 | 449.036111 | 449.036111 | 449.036111 |
| 8 | 2025-04-04 13:18:43.531508 | 1 | off | 16.990556 | 24978.543889 | 32.093272 | 29.126667 | 25.890370 | 38.835556 | 52.859506 | ... | 447.687654 | 445.260432 | 448.766420 | 435.281852 | 428.539568 | 415.594383 | 435.012160 | 425.303272 | 425.303272 | 425.303272 |
| 9 | 2025-04-04 13:18:44.565058 | 1 | off | 16.990556 | 24978.543889 | 31.014506 | 39.105247 | 51.780741 | 29.396358 | 63.107778 | ... | 433.124321 | 458.205617 | 449.845185 | 447.687654 | 442.833210 | 439.057531 | 423.145741 | 439.866605 | 439.866605 | 439.866605 |
| 10 | 2025-04-04 13:18:45.595041 | 1 | off | 16.990556 | 24978.543889 | 22.923765 | 31.553889 | 55.017037 | 56.904877 | 46.926296 | ... | 417.212531 | 438.787840 | 431.506173 | 446.069506 | 422.066975 | 419.370062 | 452.272407 | 428.539568 | 428.539568 | 428.539568 |
| 11 | 2025-04-04 13:18:46.624883 | 1 | off | 16.990556 | 24978.543889 | 32.093272 | 27.238827 | 28.047901 | 29.126667 | 47.735370 | ... | 431.506173 | 435.821235 | 436.630309 | 460.363148 | 428.809259 | 440.405988 | 453.890556 | 438.787840 | 438.787840 | 438.787840 |
| 12 | 2025-04-04 13:18:47.658044 | 1 | off | 16.990556 | 24978.543889 | 26.699444 | 39.105247 | 36.678025 | 15.642099 | 52.050432 | ... | 454.429938 | 436.360617 | 442.024136 | 430.157716 | 414.245926 | 442.293827 | 455.239012 | 451.463333 | 451.463333 | 451.463333 |
| 13 | 2025-04-04 13:18:48.687960 | 1 | off | 16.990556 | 24978.543889 | 28.856975 | 48.005062 | 20.226852 | 31.553889 | 71.198519 | ... | 450.384568 | 430.697099 | 435.281852 | 444.451358 | 427.730494 | 423.685123 | 427.191111 | 448.766420 | 448.766420 | 448.766420 |
| 14 | 2025-04-04 13:18:49.728411 | 1 | off | 16.990556 | 24978.543889 | 46.386914 | 16.720864 | 24.811605 | 49.353519 | 66.074383 | ... | 445.530123 | 414.515617 | 448.227037 | 417.482222 | 448.496728 | 453.351173 | 438.248457 | 452.002716 | 452.002716 | 452.002716 |
| 15 | 2025-04-04 13:18:50.761486 | 1 | off | 16.990556 | 24978.543889 | 43.420309 | 33.711420 | 38.835556 | 42.071852 | 33.441728 | ... | 426.112346 | 430.697099 | 435.551543 | 419.639753 | 442.293827 | 444.721049 | 445.799815 | 443.372593 | 443.372593 | 443.372593 |
| 16 | 2025-04-04 13:18:51.804446 | 1 | off | 16.990556 | 24978.543889 | 49.029889 | 31.553889 | 54.612500 | 54.855222 | 56.797000 | ... | 444.424389 | 446.366167 | 434.472778 | 437.870889 | 425.249333 | 440.055389 | 437.142722 | 441.754444 | 441.754444 | 441.754444 |
| 17 | 2025-04-04 13:18:52.839511 | 1 | off | 16.990556 | 24978.543889 | 48.274753 | 22.923765 | 25.890370 | 37.756790 | 51.241358 | ... | 440.405988 | 435.281852 | 447.417963 | 419.370062 | 424.224506 | 453.351173 | 436.900000 | 436.360617 | 436.360617 | 436.360617 |
| 18 | 2025-04-04 13:18:53.878424 | 1 | off | 16.990556 | 24978.543889 | 42.476389 | 30.097556 | 45.146333 | 61.894167 | 50.971667 | ... | 430.346500 | 416.754056 | 439.569944 | 423.793000 | 442.968056 | 466.269389 | 450.735167 | 458.259556 | 458.259556 | 458.259556 |
| 19 | 2025-04-04 13:18:54.916155 | 1 | off | 16.990556 | 24978.543889 | 25.890370 | 25.890370 | 51.780741 | 30.475123 | 61.489630 | ... | 437.978765 | 442.024136 | 432.584938 | 438.518148 | 450.654259 | 449.575494 | 460.632840 | 452.272407 | 452.272407 | 452.272407 |
| 20 | 2025-04-04 13:18:55.948033 | 1 | off | 16.990556 | 24978.543889 | 59.601790 | 38.296173 | 29.666049 | 50.162593 | 24.541914 | ... | 444.181667 | 463.599444 | 450.114877 | 422.606358 | 468.184198 | 443.102901 | 443.642284 | 439.596914 | 439.596914 | 439.596914 |
| 21 | 2025-04-04 13:18:56.982133 | 1 | off | 16.990556 | 24978.543889 | 30.744815 | 42.611235 | 33.441728 | 31.823580 | 69.040988 | ... | 418.021605 | 428.000185 | 425.572963 | 442.833210 | 442.024136 | 445.260432 | 419.639753 | 460.363148 | 460.363148 | 460.363148 |
| 22 | 2025-04-04 13:18:58.028200 | 1 | off | 16.990556 | 24978.543889 | 27.427611 | 38.835556 | 20.145944 | 31.553889 | 34.466556 | ... | 440.298111 | 455.589611 | 441.269000 | 441.026278 | 425.006611 | 456.803222 | 439.569944 | 443.938944 | 443.938944 | 443.938944 |
| 23 | 2025-04-04 13:18:59.065314 | 1 | off | 16.990556 | 24978.543889 | 46.117222 | 22.654074 | 29.666049 | 25.890370 | 47.195988 | ... | 450.923951 | 437.169691 | 430.966790 | 448.496728 | 440.405988 | 447.417963 | 440.945370 | 438.248457 | 438.248457 | 438.248457 |
| 24 | 2025-04-04 13:19:00.095774 | 1 | off | 16.990556 | 24978.543889 | 27.427611 | 19.175056 | 21.845000 | 35.437444 | 56.311556 | ... | 459.230444 | 430.346500 | 448.550667 | 450.007000 | 441.511722 | 440.055389 | 448.065222 | 437.628167 | 437.628167 | 437.628167 |
| 25 | 2025-04-04 13:19:01.128261 | 1 | off | 16.990556 | 24978.543889 | 37.217407 | 14.293642 | 28.317593 | 41.532469 | 47.735370 | ... | 436.090926 | 438.518148 | 440.405988 | 449.575494 | 436.630309 | 434.472778 | 456.857160 | 436.900000 | 436.900000 | 436.900000 |
| 26 | 2025-04-04 13:19:02.160391 | 1 | off | 16.990556 | 24978.543889 | 38.350111 | 26.942167 | 51.699833 | 39.321000 | 55.097944 | ... | 447.579778 | 415.055000 | 426.220222 | 448.065222 | 453.890556 | 439.084500 | 436.171833 | 442.968056 | 442.968056 | 442.968056 |
| 27 | 2025-04-04 13:19:03.202006 | 1 | off | 16.990556 | 24978.543889 | 17.529938 | 40.993086 | 41.532469 | 46.926296 | 44.499074 | ... | 450.114877 | 456.048086 | 430.427407 | 442.563519 | 452.002716 | 427.460802 | 436.090926 | 438.248457 | 438.248457 | 438.248457 |
| 28 | 2025-04-04 13:19:04.242210 | 1 | off | 16.990556 | 24978.543889 | 40.777333 | 30.583000 | 21.845000 | 37.864667 | 57.767889 | ... | 445.395278 | 432.045556 | 443.210778 | 457.531389 | 459.230444 | 443.453500 | 444.181667 | 436.414556 | 436.414556 | 436.414556 |
| 29 | 2025-04-04 13:19:05.280109 | 1 | off | 16.990556 | 24978.543889 | 39.914321 | 42.341543 | 25.890370 | 47.735370 | 55.286728 | ... | 420.988210 | 461.981296 | 433.933395 | 445.530123 | 432.315247 | 430.966790 | 468.184198 | 435.551543 | 435.551543 | 435.551543 |
| 30 | 2025-04-04 13:19:06.334964 | 1 | off | 16.990556 | 24978.543889 | 47.330833 | 22.815889 | 44.418167 | 56.797000 | 54.127056 | ... | 436.171833 | 445.395278 | 470.152944 | 423.550278 | 433.744611 | 440.055389 | 435.686389 | 432.288278 | 432.288278 | 432.288278 |
| 31 | 2025-04-04 13:19:07.367506 | 1 | off | 16.990556 | 24978.543889 | 37.217407 | 27.778210 | 31.553889 | 29.935741 | 46.117222 | ... | 450.654259 | 430.427407 | 424.763889 | 450.654259 | 428.539568 | 447.957346 | 458.205617 | 414.515617 | 414.515617 | 414.515617 |
| 32 | 2025-04-04 13:19:08.397854 | 1 | off | 16.990556 | 24978.543889 | 19.417778 | 21.602278 | 53.156167 | 47.816278 | 58.738778 | ... | 425.734778 | 431.074667 | 465.055778 | 443.453500 | 457.774111 | 450.007000 | 429.375611 | 440.055389 | 440.055389 | 440.055389 |
| 33 | 2025-04-04 13:19:09.427097 | 1 | off | 16.990556 | 24978.543889 | 31.014506 | 16.990556 | 30.475123 | 34.250802 | 61.759321 | ... | 450.923951 | 466.835741 | 455.778395 | 454.969321 | 448.496728 | 444.181667 | 456.048086 | 430.697099 | 430.697099 | 430.697099 |
| 34 | 2025-04-04 13:19:10.459716 | 1 | off | 16.990556 | 24978.543889 | 42.880926 | 25.350988 | 49.353519 | 52.859506 | 56.904877 | ... | 453.620864 | 463.329753 | 446.608889 | 450.384568 | 426.382037 | 446.878580 | 425.033580 | 424.763889 | 424.763889 | 424.763889 |
| 35 | 2025-04-04 13:19:11.491974 | 1 | off | 16.990556 | 24978.543889 | 48.274753 | 23.732840 | 23.193457 | 43.420309 | 68.231914 | ... | 464.138827 | 446.878580 | 434.472778 | 427.191111 | 429.348642 | 449.575494 | 443.911975 | 432.045556 | 432.045556 | 432.045556 |
| 36 | 2025-04-04 13:19:12.537711 | 1 | off | 16.990556 | 24978.543889 | 26.429753 | 11.866420 | 58.253333 | 47.195988 | 54.477654 | ... | 428.539568 | 410.200556 | 443.911975 | 435.281852 | 435.281852 | 451.193642 | 427.460802 | 444.990741 | 444.990741 | 444.990741 |
| 37 | 2025-04-04 13:19:13.572105 | 1 | off | 16.990556 | 24978.543889 | 39.105247 | 12.405802 | 27.508519 | 33.711420 | 52.050432 | ... | 438.787840 | 444.721049 | 435.551543 | 450.384568 | 452.811790 | 440.405988 | 443.911975 | 448.496728 | 448.496728 | 448.496728 |
| 38 | 2025-04-04 13:19:14.604002 | 1 | off | 16.990556 | 24978.543889 | 28.047901 | 20.766235 | 52.589815 | 32.902346 | 50.971667 | ... | 430.157716 | 430.966790 | 440.675679 | 451.463333 | 422.336667 | 423.145741 | 442.293827 | 432.045556 | 432.045556 | 432.045556 |
| 39 | 2025-04-04 13:19:15.637091 | 1 | off | 16.990556 | 24978.543889 | 24.272222 | 48.544444 | 36.947716 | 49.623210 | 57.174568 | ... | 441.754444 | 445.260432 | 443.372593 | 440.136296 | 447.417963 | 464.408519 | 452.002716 | 450.654259 | 450.654259 | 450.654259 |
| 40 | 2025-04-04 13:19:16.672483 | 1 | off | 16.990556 | 24978.543889 | 30.744815 | 46.656605 | 33.172037 | 44.229383 | 35.059877 | ... | 442.024136 | 443.911975 | 449.305802 | 473.578025 | 440.945370 | 427.730494 | 444.990741 | 448.496728 | 448.496728 | 448.496728 |
| 41 | 2025-04-04 13:19:17.706073 | 1 | off | 16.990556 | 24978.543889 | 26.699444 | 25.485833 | 38.835556 | 39.563722 | 34.223833 | ... | 427.919278 | 464.570333 | 442.968056 | 430.589222 | 443.210778 | 447.822500 | 436.414556 | 434.230056 | 434.230056 | 434.230056 |
| 42 | 2025-04-04 13:19:18.742677 | 1 | off | 16.990556 | 24978.543889 | 42.341543 | 25.350988 | 37.217407 | 48.814136 | 53.398889 | ... | 433.663704 | 434.742469 | 429.618333 | 416.673148 | 452.002716 | 450.384568 | 427.191111 | 437.709074 | 437.709074 | 437.709074 |
| 43 | 2025-04-04 13:19:19.775702 | 1 | off | 16.990556 | 24978.543889 | 19.957160 | 19.148086 | 30.205432 | 35.059877 | 60.950247 | ... | 436.630309 | 451.733025 | 443.372593 | 444.721049 | 439.057531 | 449.575494 | 452.542099 | 426.382037 | 426.382037 | 426.382037 |
| 44 | 2025-04-04 13:19:20.805906 | 1 | off | 16.990556 | 24978.543889 | 28.856975 | 18.608704 | 24.811605 | 34.250802 | 48.274753 | ... | 453.890556 | 457.126852 | 443.102901 | 454.429938 | 430.427407 | 456.587469 | 459.014691 | 438.248457 | 438.248457 | 438.248457 |
| 45 | 2025-04-04 13:19:21.836857 | 1 | off | 16.990556 | 24978.543889 | 39.374938 | 45.308148 | 22.923765 | 46.926296 | 51.780741 | ... | 432.854630 | 438.248457 | 461.981296 | 421.257901 | 444.990741 | 424.763889 | 445.260432 | 432.854630 | 432.854630 | 432.854630 |
| 46 | 2025-04-04 13:19:22.871144 | 1 | off | 16.990556 | 24978.543889 | 46.117222 | 30.825722 | 21.359556 | 50.971667 | 54.612500 | ... | 439.812667 | 450.007000 | 443.210778 | 427.919278 | 446.123444 | 436.657278 | 435.686389 | 438.113611 | 438.113611 | 438.113611 |
| 47 | 2025-04-04 13:19:23.908302 | 1 | off | 16.990556 | 24978.543889 | 36.678025 | 40.184012 | 36.678025 | 38.026481 | 49.623210 | ... | 456.857160 | 446.339198 | 452.002716 | 449.036111 | 438.248457 | 430.966790 | 444.990741 | 429.078951 | 429.078951 | 429.078951 |
| 48 | 2025-04-04 13:19:24.941178 | 1 | off | 16.990556 | 24978.543889 | 42.476389 | 23.058611 | 19.417778 | 60.437833 | 46.359944 | ... | 444.181667 | 419.666722 | 431.074667 | 431.802833 | 450.977889 | 439.327222 | 444.667111 | 449.764278 | 449.764278 | 449.764278 |
| 49 | 2025-04-04 13:19:25.976058 | 1 | off | 16.990556 | 24978.543889 | 51.511049 | 24.272222 | 33.711420 | 52.320123 | 46.656605 | ... | 439.327222 | 427.191111 | 442.293827 | 436.090926 | 438.787840 | 457.396543 | 440.945370 | 476.005247 | 476.005247 | 476.005247 |
| 50 | 2025-04-04 13:19:27.008444 | 1 | off | 16.990556 | 24978.543889 | 39.105247 | 31.823580 | 39.374938 | 48.544444 | 43.959691 | ... | 465.756975 | 429.888025 | 436.090926 | 446.069506 | 452.811790 | 452.272407 | 422.606358 | 450.923951 | 450.923951 | 450.923951 |
| 51 | 2025-04-04 13:19:28.045717 | 1 | off | 16.990556 | 24978.543889 | 49.892901 | 51.511049 | 49.083827 | 28.856975 | 68.501605 | ... | 464.678210 | 440.136296 | 429.618333 | 453.081481 | 443.372593 | 436.900000 | 429.078951 | 444.451358 | 444.451358 | 444.451358 |
| 52 | 2025-04-04 13:19:29.076808 | 1 | off | 16.990556 | 24978.543889 | 31.553889 | 12.405802 | 43.690000 | 38.565864 | 67.153148 | ... | 446.069506 | 463.869136 | 433.933395 | 441.754444 | 440.136296 | 431.775864 | 449.575494 | 435.012160 | 435.012160 | 435.012160 |
| 53 | 2025-04-04 13:19:30.113538 | 1 | off | 16.990556 | 24978.543889 | 45.308148 | 30.475123 | 34.520494 | 14.023951 | 46.117222 | ... | 460.632840 | 444.181667 | 438.248457 | 440.405988 | 437.978765 | 444.451358 | 427.191111 | 440.405988 | 440.405988 | 440.405988 |
| 54 | 2025-04-04 13:19:31.146010 | 1 | off | 16.990556 | 24978.543889 | 54.207963 | 27.508519 | 37.217407 | 35.599259 | 51.780741 | ... | 447.687654 | 442.024136 | 470.341728 | 459.554074 | 424.494198 | 434.742469 | 444.181667 | 436.360617 | 436.360617 | 436.360617 |
| 55 | 2025-04-04 13:19:32.180604 | 1 | off | 16.990556 | 24978.543889 | 30.475123 | 33.981111 | 43.690000 | 40.993086 | 52.050432 | ... | 429.348642 | 444.990741 | 419.909444 | 431.506173 | 430.427407 | 437.709074 | 461.981296 | 444.181667 | 444.181667 | 444.181667 |
| 56 | 2025-04-04 13:19:33.213159 | 1 | off | 16.990556 | 24978.543889 | 35.329568 | 29.666049 | 16.990556 | 37.217407 | 57.983642 | ... | 433.124321 | 441.484753 | 441.484753 | 455.778395 | 451.463333 | 437.439383 | 442.563519 | 438.787840 | 438.787840 | 438.787840 |
| 57 | 2025-04-04 13:19:46.247957 | 1 | static | 16.990556 | 24978.543889 | 42.840472 | 38.350111 | 28.277139 | 32.767500 | 58.010611 | ... | 426.220222 | 443.817583 | 446.123444 | 449.036111 | 430.467861 | 437.264083 | 419.545361 | 451.220611 | 451.220611 | 451.220611 |
58 rows × 2051 columns
import numpy as np
intensities=np.array(exp_data[(exp_data["type"] != "zero") & (exp_data["type"] != "static")].iloc[:, 3:], dtype=np.float64)
static=np.array(exp_data[(exp_data["type"]== "static")].iloc[:, 3:], dtype=np.float64)[0]
zero=np.array(exp_data[(exp_data["type"]== "zero")].iloc[:, 3:], dtype=np.float64)[0]
wavelengths = np.array(exp_data.columns[3:], dtype=np.float64)
timestamps = pd.to_datetime(exp_data["timestamp"][(exp_data["type"] != "zero") & (exp_data["type"] != "static")]) # Convert timestamp strings to datetime objects
timestamps = np.array((timestamps - timestamps.iloc[0]).dt.total_seconds()) # Convert to seconds since the first timestamp
def compute_absorbance(intensities: np.ndarray, static: np.ndarray, zero: np.ndarray) -> np.ndarray:
EPS = 1e-12
num = intensities - static
den = np.maximum(zero - static, EPS) # Éviter division par zéro
absorbance = -np.log10(np.maximum(num / den, EPS)) # Éviter log(0) ou log(négatif)
return absorbance
absorbance = compute_absorbance(intensities, static, zero)
Show code cell source
import matplotlib.pyplot as plt
import matplotlib.cm as cm
# Set global style
plt.style.use("ggplot") # Clean style
fig, axs = plt.subplots(1, 3, figsize=(18, 5))
fig.suptitle("Spectral Analysis", fontsize=16)
# --- Intensity Plot ---
axs[0].plot(wavelengths, intensities[0, :], label=f"I at t = {timestamps[0]:.0f} s", linewidth=2)
axs[0].plot(wavelengths, intensities[len(intensities)//2, :], label=f"I at t = {timestamps[len(intensities)//2]:.0f} s", linewidth=2)
axs[0].plot(wavelengths, intensities[-1, :], label=f"I at t = {timestamps[-1]:.0f} s", linewidth=2)
axs[0].plot(wavelengths, static, '--', label="Static", linewidth=2)
axs[0].plot(wavelengths, zero, '--', label="Zero", linewidth=2)
axs[0].set_title("Intensity over Wavelength")
axs[0].set_xlabel("Wavelength (nm)")
axs[0].set_ylabel("Intensity")
axs[0].legend()
axs[0].grid(True, which='major', linestyle='--', linewidth=0.6, color='gray', alpha=0.5, zorder=0)
axs[0].minorticks_on()
axs[0].grid(True, which='minor', linestyle=':', linewidth=0.3, color='lightgray', alpha=0.4, zorder=0)
# --- Absorbance Plot ---
axs[1].plot(wavelengths, absorbance[0, :], label=f"t = {timestamps[0]:.0f} s", linewidth=2)
axs[1].plot(wavelengths, absorbance[len(absorbance)//2, :], label=f"t = {timestamps[len(absorbance)//2]:.0f} s", linewidth=2)
axs[1].plot(wavelengths, absorbance[-1, :], label=f"t = {timestamps[-1]:.0f} s", linewidth=2)
axs[1].set_title("Absorbance over Wavelength")
axs[1].set_xlabel("Wavelength (nm)")
axs[1].set_ylabel("Absorbance")
axs[1].set_ylim(-0.4, 2)
axs[1].legend()
axs[1].grid(True, which='major', linestyle='--', linewidth=0.6, color='gray', alpha=0.5, zorder=0)
axs[1].minorticks_on()
axs[1].grid(True, which='minor', linestyle=':', linewidth=0.3, color='lightgray', alpha=0.4, zorder=0)
# --- Absorbance vs Time Plot ---
WL = [482, 408]
idxs = [np.argmin(np.abs(wavelengths - wl)) for wl in WL]
for i, idx in enumerate(idxs):
axs[2].plot(timestamps, absorbance[:, idx], label=f"{wavelengths[idx]:.0f} nm", linewidth=2)
axs[2].set_title("Absorbance over Time")
axs[2].set_xlabel("Time (s)")
axs[2].set_ylabel("Absorbance")
axs[2].legend()
axs[2].grid(True, which='major', linestyle='--', linewidth=0.6, color='gray', alpha=0.5, zorder=0)
axs[2].minorticks_on()
axs[2].grid(True, which='minor', linestyle=':', linewidth=0.3, color='lightgray', alpha=0.4, zorder=0)
plt.tight_layout(rect=[0, 0, 1, 0.95])
plt.show()
# la premiere mesure doit etre viré
# Define wavelength range of interest
wavelength_range = [480, 484]
idx_range = np.argmin(np.abs(wavelengths[:, None] - wavelength_range), axis=0)
# Extract and normalize absorbance data within the wavelength range
absorbance_subset = absorbance[:, idx_range[0]:idx_range[1]]
# absorbance_subset -= np.min(absorbance_subset)
wavelengths_subset = wavelengths[idx_range[0]:idx_range[1]]
# Filter out specific time points (e.g., first frame and last 300 entries)
# Adjust indices as needed
time_filter_indices = [0] #+ list(range(-45, 0))
absorbance_subset = np.delete(absorbance_subset, time_filter_indices, axis=0)
timestamps_filtered = np.delete(timestamps, time_filter_indices)
# Create subplots
fig, axs = plt.subplots(1, 3, figsize=(18, 5))
# --- 1. Absorbance Heatmap ---
X, Y = np.meshgrid(timestamps_filtered, wavelengths_subset)
contour = axs[0].contourf(X, Y, absorbance_subset.T, levels=30, cmap="coolwarm")
axs[0].set_title("Experimental Absorbance Data")
axs[0].set_xlabel("Time (s)")
axs[0].set_ylabel("Wavelength (nm)")
fig.colorbar(contour, ax=axs[0])
# --- 2. Absorbance Over Time (Averaged Across Wavelength Range) ---
mean_absorbance = np.mean(absorbance_subset, axis=1)
axs[1].plot(timestamps_filtered, mean_absorbance, label=f"Mean absorbance ({wavelengths_subset[0]:.0f}-{wavelengths_subset[-1]:.0f} nm)", color='tab:blue')
axs[1].set_title("Mean Absorbance Over Time")
axs[1].set_xlabel("Time (s)")
axs[1].set_ylabel("Absorbance")
axs[1].tick_params(axis="x", rotation=45)
axs[1].legend()
axs[1].grid(True)
# --- 3. Absorbance Spectrum at Start and End ---
axs[2].plot(wavelengths_subset, absorbance_subset[0, :], label="Spectrum at $t=0$", color='red')
axs[2].plot(wavelengths_subset, absorbance_subset[-1, :], label="Spectrum at $t=end$", color='blue')
axs[2].set_title("Absorbance Spectra")
axs[2].set_xlabel("Wavelength (nm)")
axs[2].set_ylabel("Absorbance")
axs[2].legend()
axs[2].grid(True)
plt.tight_layout()
plt.show()
from scipy.integrate import solve_ivp
from scipy.optimize import curve_fit
import numpy as np
import matplotlib.pyplot as plt
# === Physics constants ===
h = 6.62607004 * 10 ** (-34) # Planck's constant (J·s)
NA = 6.02214086 * 10 ** (23) # Avogadro's number (mol^-1)
c_vaccum = 299792458 # Speed of light in vacuum (m/s)
wl = 405 # Wavelength (nm)
v = c_vaccum / (wl * 1e-9) # Frequency (Hz)
volume = 23e-4 # Volume (L)
l = 1 # Path length (cm)
eps_on_482 = 65474
eps_off_482 = 65
eps_on_405 = 0.1847826087*eps_on_482
eps_off_405 = 25343
I_w_list = [ 3.8e-3 ] # No actinometry for 405nm LED
# === Experimental data ===
# Assuming 'timestamps_filtered' and 'C_exp' are already defined
# === Initialize parameters ===
lpopt = []
for I_w in I_w_list:
# Compute initial concentration based on experimental data
C_exp = mean_absorbance / (eps_on_482 * l)
C0 = C_exp[-1]
I_0 = I_w / (h * v * NA) / volume # Photon irradiance (mol·L^-1·s^-1)
# === ODE definition ===
def dC_dt(t, C_ON, QY_ON2OFF, QY_OFF2ON):
C_OFF = C0 - C_ON
Iabs_ON = I_0 * (1 - np.exp(-eps_on_405 * C_ON * l * np.log(10)))
Iabs_OFF = I_0 * (1 - np.exp(-eps_on_405 * C_OFF * l * np.log(10)))
dCCF_deri = -QY_ON2OFF * Iabs_ON + QY_OFF2ON * Iabs_OFF
return dCCF_deri
# === Fitting function wrapper ===
def model(t, QY_ON2OFF, QY_OFF2ON, offset):
sol = solve_ivp(dC_dt, [t[0], t[-1]], [C_exp[0]], args=(QY_ON2OFF, QY_OFF2ON), t_eval=t)
if not sol.success:
print("⚠️ ODE solver failed:", sol.message)
return np.full_like(t, np.nan)
return sol.y[0]+ offset
# === Fit the model to the experimental data ===
initial_guess = (0.001, 0.2, 0) # QY_ON2OFF, QY_OFF2ON, offset
bounds = ([1e-4, 1e-4,-1], [0.1, 1,1]) # ([min1, min2, ...], [max1, max2, ...])
popt, pcov = curve_fit(model, timestamps_filtered, C_exp, p0=initial_guess, bounds=bounds)
# Extract fitted parameters and compute uncertainties
uncertainties = np.sqrt(np.diag(pcov))
lpopt.append(popt)
print(f"=== Fit Results I={I_w:0.3e} W ===")
for i, (coef, incert) in enumerate(zip(popt, uncertainties), start=1):
print(f"Parameter {i}: {coef:.4f} ± {incert:.4f}")
# Compute fitted values
fitted_values = model(timestamps_filtered, *popt)
# === Plotting ===
fig, a = plt.subplots(1, 2, figsize=(15, 5))
pastel_colors = ["m", "#f9955e", "#5080e0"] # Mauve, orange, sky blue
# First Plot - Concentration Over Time
a[0].plot(timestamps_filtered, C_exp, "1", color=pastel_colors[2], label="C exp", alpha=0.6)
a[0].plot(timestamps_filtered, fitted_values, "-", color=pastel_colors[0], label=f"C model for QY1={popt[0]:0.4f}, QY2={popt[1]:0.4f}")
a[0].set_xlabel("Time (s)", fontsize=12, fontweight="bold")
a[0].set_ylabel("Concentration (M)", fontsize=12, fontweight="bold")
a[0].set_title("Concentration Over Time", fontsize=14, fontweight="bold")
a[0].tick_params(axis="x", rotation=45)
a[0].grid(linestyle="--", alpha=0.5)
a[0].legend()
# Second Plot - Residuals Over Time
residuals = C_exp - fitted_values
a[1].plot(timestamps_filtered, residuals, "1", color=pastel_colors[0], label="Residuals", alpha=0.9)
a[1].set_xlabel("Time (s)", fontsize=12, fontweight="bold")
a[1].set_ylabel("Residuals", fontsize=12, fontweight="bold")
a[1].set_title("Residuals", fontsize=14, fontweight="bold")
a[1].tick_params(axis="x", rotation=45)
a[1].grid(linestyle="--", alpha=0.5)
a[1].legend()
plt.tight_layout()
plt.show()
=== Fit Results I=3.800e-03 W ===
Parameter 1: 0.0014 ± 0.0013
Parameter 2: 0.4951 ± 0.0044
Parameter 3: 0.0000 ± 0.0000