Photofatigue Measurements#
1. Import Data and Preprocess for Photofatigue Measurements#
In this first step, we import the necessary experimental data from a CSV file.
Timestamp filtering: Only data after
2025-04-08is used for analysis.
import pandas as pd
# Load experimental data
exp_data = pd.read_csv("DATA/Spectrumsfatigue.csv")
# Filter data to include only entries after a specific timestamp
exp_data = exp_data[exp_data["timestamp"] > "2025-04-08"]
# Display the filtered dataset
exp_data
| timestamp | cycle | type | 186.85486 | 187.31995223015844 | 187.78500323297956 | 188.250012996982 | 188.71498151068454 | 189.17990876260572 | 189.64479474126426 | ... | 1032.9632529736894 | 1033.3204920667242 | 1033.6776665334785 | 1034.034776362471 | 1034.3918215422202 | 1034.7488020612445 | 1035.1057179080635 | 1035.4625690711953 | 1035.8193555391586 | 1036.176077300472 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 863 | 2025-04-08 09:38:38.241780 | 0 | zero | 16.990556 | 24978.543889 | 347.456861 | 334.956667 | 350.490889 | 359.228889 | 371.365000 | ... | 759.235111 | 750.739833 | 755.958361 | 750.497111 | 755.715639 | 744.793139 | 754.866111 | 767.123583 | 767.123583 | 767.123583 |
| 864 | 2025-04-08 09:39:44.486116 | 1 | on | 16.990556 | 24978.543889 | 352.796750 | 356.680306 | 352.796750 | 362.262917 | 357.772556 | ... | 746.977639 | 739.453250 | 747.220361 | 749.162139 | 757.050611 | 759.720556 | 766.516778 | 749.768944 | 749.768944 | 749.768944 |
| 865 | 2025-04-08 09:40:19.529467 | 1 | off | 16.990556 | 24978.543889 | 358.986167 | 352.554028 | 341.631528 | 371.365000 | 379.010750 | ... | 750.133028 | 751.468000 | 743.458167 | 763.968194 | 746.128111 | 754.866111 | 750.254389 | 757.414694 | 757.414694 | 757.414694 |
| 866 | 2025-04-08 09:40:25.564251 | 1 | static | 16.990556 | 24978.543889 | 357.408472 | 358.500722 | 346.850056 | 365.418306 | 381.802056 | ... | 757.293333 | 741.273667 | 763.361389 | 770.643056 | 762.754583 | 767.244944 | 753.773861 | 760.327361 | 760.327361 | 760.327361 |
| 867 | 2025-04-08 09:40:40.608372 | 2 | on | 16.990556 | 24978.543889 | 365.054222 | 348.063667 | 360.199778 | 361.170667 | 372.093167 | ... | 748.676694 | 744.914500 | 757.293333 | 764.089556 | 756.079722 | 753.167056 | 758.992389 | 746.249472 | 746.249472 | 746.249472 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 1546 | 2025-04-08 13:12:50.290246 | 228 | off | 16.990556 | 24978.543889 | 133.861306 | 130.705917 | 121.361111 | 138.230306 | 137.987583 | ... | 527.192667 | 531.561667 | 525.979056 | 522.823667 | 544.425944 | 529.255806 | 546.489083 | 529.741250 | 529.741250 | 529.741250 |
| 1547 | 2025-04-08 13:12:56.349038 | 228 | static | 16.990556 | 24978.543889 | 114.928972 | 119.540694 | 125.001944 | 127.065083 | 139.686639 | ... | 534.595694 | 513.721583 | 545.032750 | 522.459583 | 520.153722 | 537.629722 | 546.246361 | 533.867528 | 533.867528 | 533.867528 |
| 1548 | 2025-04-08 13:13:11.473565 | 229 | on | 16.990556 | 24978.543889 | 118.448444 | 124.880583 | 126.094194 | 121.482472 | 145.026528 | ... | 532.047111 | 529.377167 | 523.915917 | 534.959778 | 522.338222 | 530.712139 | 535.930667 | 543.455056 | 543.455056 | 543.455056 |
| 1549 | 2025-04-08 13:13:46.542295 | 229 | off | 16.990556 | 24978.543889 | 122.332000 | 109.103639 | 114.564889 | 131.070000 | 147.332389 | ... | 528.527639 | 536.658833 | 526.221778 | 529.983972 | 530.712139 | 527.678111 | 536.294750 | 537.022917 | 537.022917 | 537.022917 |
| 1550 | 2025-04-08 13:13:52.603917 | 229 | static | 16.990556 | 24978.543889 | 117.113472 | 109.710444 | 104.491917 | 120.754306 | 151.580028 | ... | 547.824056 | 526.585861 | 545.275472 | 530.590778 | 553.285306 | 511.051639 | 528.891722 | 517.241056 | 517.241056 | 517.241056 |
688 rows × 2051 columns
2. Data Preparation: Intensity Arrays & Absorbance Calculation#
Now, we prepare the intensity data based on the “on”, “off”, “static”, and “zero” conditions. These intensity values will be used to compute the absorbance values.
Intensity arrays: Data for “on” and “off” states are extracted, along with the static and zero intensity references.
Absorbance formula: Absorbance is calculated from the intensity data using the formula:
$$ A = -\log_{10}\left(\frac{I_{\text{on}} - I_{\text{static}}}{I_{\text{zero}} - I_{\text{static}}}\right) $$
where:
$I_{\text{on}}$ is the intensity during the “on” state,
$I_{\text{static}}$ is the static reference intensity,
$I_{\text{zero}}$ is the zero intensity reference.
import numpy as np
# Extract relevant data for 'on', 'off', 'static', and 'zero' conditions
I_on = np.array(exp_data[exp_data["type"] == "on"].iloc[:, 3:], dtype=np.float64)
I_off = np.array(exp_data[exp_data["type"] == "off"].iloc[:, 3:], dtype=np.float64)
static = np.array(exp_data[(exp_data["type"] == "static")].iloc[:, 3:], dtype=np.float64)
zero = np.array(exp_data[(exp_data["type"] == "zero")].iloc[:, 3:], dtype=np.float64)
# Extract wavelengths (assumed to be in columns from 3 onward)
wavelengths = np.array(exp_data.columns[3:], dtype=np.float64)
# Convert timestamps to seconds since the first "on" timestamp
t_on = pd.to_datetime(exp_data["timestamp"][exp_data["type"] == "on"])
t_on = np.array((t_on - t_on.iloc[0]).dt.total_seconds())
t_off = pd.to_datetime(exp_data["timestamp"][exp_data["type"] == "off"])
t_off = np.array((t_off - t_off.iloc[0]).dt.total_seconds())
# Function to calculate absorbance
def compute_absorbance(intensities: np.ndarray, static: np.ndarray, zero: np.ndarray) -> np.ndarray:
EPS = 1e-12 # Small epsilon to avoid division by zero
num = intensities - static
den = np.maximum(zero - static, EPS) # Prevent division by zero
absorbance = -np.log10(np.maximum(num / den, EPS)) # Avoid log(0) or negative
return absorbance
# Calculate absorbance for "on" and "off" intensities
A_on = compute_absorbance(I_on, static, zero)
A_off = compute_absorbance(I_off, static, zero)
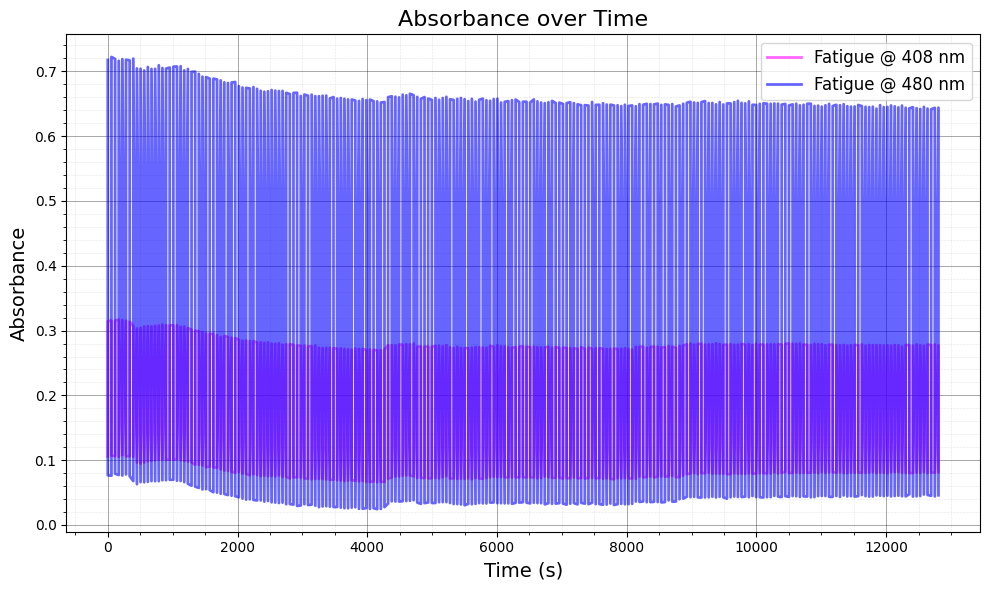
3. Plot Absorbance vs Time for Selected Wavelengths#
In this section, we visualize the absorbance data over time for selected wavelengths (408 nm and 480 nm). This helps to assess the photofatigue effects at these specific wavelengths.
Plotting: We plot the absorbance for both “on” and “off” conditions for each of the selected wavelengths.
The absorbance is shown as a function of time for the given wavelength range.
import matplotlib.pyplot as plt
# Define target wavelengths (in nm)
wavelength_target = [408, 480]
wavelength_idxs = np.argmin(np.abs(wavelengths[:, None] - np.array(wavelength_target)), axis=0)
# Create the figure and axis
fig, ax = plt.subplots(figsize=(10, 6))
colors = [["#FF00FF", "#412544"], ["blue", "red"]] # Colors for different wavelengths
# Plot absorbance over time for selected wavelengths
for i, idx in enumerate(wavelength_idxs):
t_combined = np.concatenate([t_on, t_off]) # Combine on and off times
A_combined = np.concatenate([A_on[:, idx], A_off[:, idx]]) # Combine on and off absorbance
# Sort by time
sorted_indices = np.argsort(t_combined)
t_sorted = t_combined[sorted_indices]
A_sorted = A_combined[sorted_indices]
# Plot combined data
ax.plot(
t_sorted, A_sorted,
label=f"Fatigue @ {wavelengths[idx]:.0f} nm",
color=colors[i][0], # Color for each wavelength
linewidth=2,
alpha=0.6,
)
# Set axis labels and title
ax.set_title("Absorbance over Time", fontsize=16)
ax.set_xlabel("Time (s)", fontsize=14)
ax.set_ylabel("Absorbance", fontsize=14)
# Add legend and grid
ax.legend(fontsize=12)
ax.minorticks_on()
ax.grid(which='major', linestyle='-', linewidth=0.6, color='black', alpha=0.4)
ax.grid(which='minor', linestyle=':', linewidth=0.4, color='black', alpha=0.2)
# Ensure layout is tidy
ax.set_axisbelow(True)
plt.tight_layout()
plt.show()
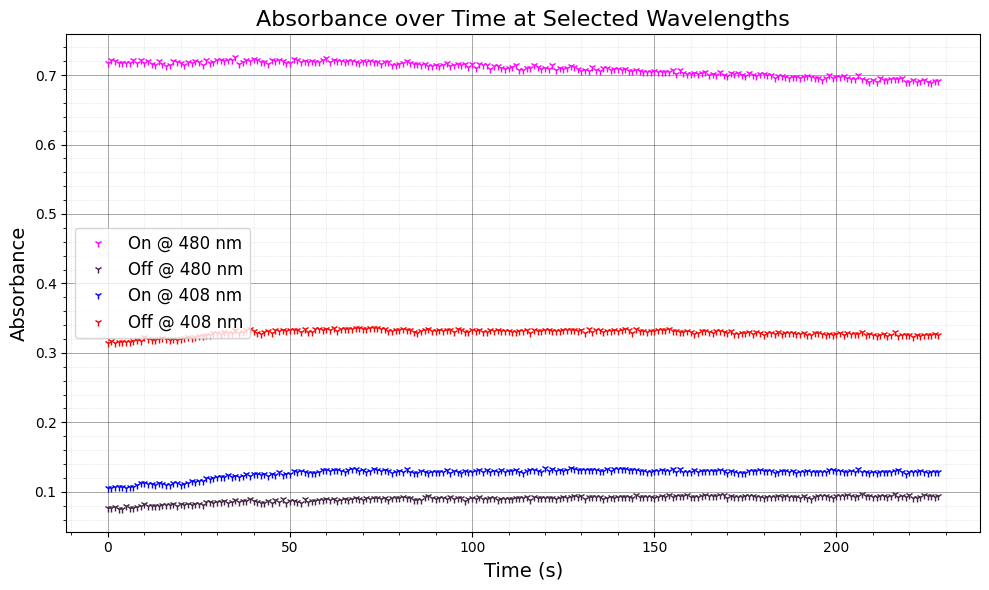
4. Adjust Absorbance for Offset and Plot#
Here, we adjust the absorbance data for the “on” and “off” conditions by subtracting the absorbance at a reference wavelength (550 nm) to account for baseline shifts. This correction ensures a more accurate comparison between wavelengths.
# Define target wavelengths
wavelength_target = [480, 408]
wavelength_idxs = np.argmin(np.abs(wavelengths[:, None] - np.array(wavelength_target)), axis=0)
# Reference wavelength for adjustment (e.g., 550 nm)
wlt = 550
id2 = np.argmin(np.abs(wavelengths[:, None] - np.array(wlt)))
# Create the figure and axis
fig, ax = plt.subplots(figsize=(10, 6))
colors = [["#FF00FF", "#412544"], ["blue", "red"]] # Colors for different wavelengths
# Plot absorbance over time with adjustments
for i, idx in enumerate(wavelength_idxs):
# Adjust "on" absorbance data
ax.plot(
A_on[:, idx] - 1 * (A_on[:, id2] - np.min(A_on[0, id2])), "1",
label=f"On @ {wavelengths[idx]:.0f} nm",
color=colors[i][0],
linewidth=2
)
# Adjust "off" absorbance data
ax.plot(
A_off[:, idx] - 1 * (A_off[:, id2] - np.min(A_off[0, id2])), "1",
label=f"Off @ {wavelengths[idx]:.0f} nm",
color=colors[i][1],
linewidth=2
)
# Set axis labels and title
ax.set_title("Absorbance over Time at Selected Wavelengths", fontsize=16)
ax.set_xlabel("Time (s)", fontsize=14)
ax.set_ylabel("Absorbance", fontsize=14)
# Add legend and grid
ax.legend(fontsize=12)
ax.minorticks_on()
ax.grid(which='major', linestyle='-', linewidth=0.6, color='black', alpha=0.4)
ax.grid(which='minor', linestyle=':', linewidth=0.4, color='black', alpha=0.2)
# Ensure layout is tidy
ax.set_axisbelow(True)
plt.tight_layout()
plt.show()
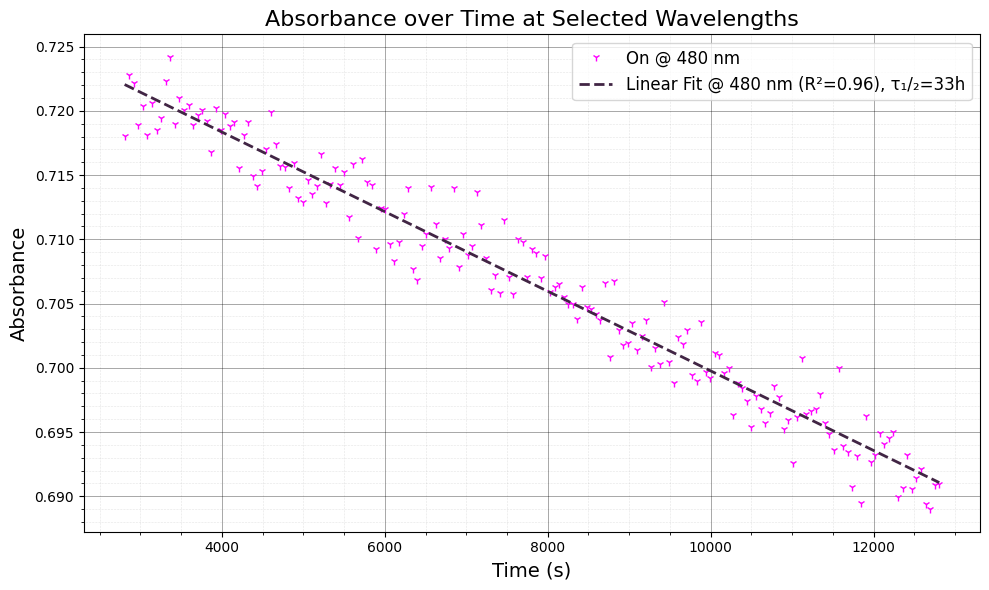
5. Linear Fit and Time Constant Estimation#
We now perform a linear regression on the absorbance data to estimate the time constant (τ) for the photofatigue process. This is calculated by fitting the absorbance data to a linear model, where the slope gives us the decay rate.
Linear regression: We fit the absorbance data after the initial part of the signal (after 50 seconds) to a linear model.
Time constant: From the linear regression results, we calculate the time constant as $( \tau_{1/2} = \frac{-\text{intercept}}{2 \times \text{slope}} )$, and express it in hours.
The time constant helps to quantify the rate of photofatigue.
import numpy as np
import matplotlib.pyplot as plt
from scipy.stats import linregress
# Define target wavelength (480 nm)
wavelength_target = [480]
wavelength_idxs = np.argmin(np.abs(wavelengths[:, None] - np.array(wavelength_target)), axis=0)
# Create the figure and axis
fig, ax = plt.subplots(figsize=(10, 6))
colors = [["#FF00FF", "#412544"], ["blue", "red"]] # Colors for different wavelengths
# Compute adjusted absorbance data
dat = A_on[:, idx] - 1 * (A_on[:, id2] - np.min(A_on[0, id2]))
# Plot absorbance data with linear regression
for i, idx in enumerate(wavelength_idxs):
t = t_on[50:] # Start from second 50 onward
y = dat[50:]
# Plot raw absorbance data
ax.plot(t, y, "1", label=f"On @ {wavelengths[idx]:.0f} nm", color=colors[i][0], linewidth=2)
# Perform linear regression
slope, intercept, r_value, p_value, std_err = linregress(t, y)
y_fit = slope * t + intercept
# Plot regression line and annotate with R^2 and time constant
ax.plot(t, y_fit, "--", label=f"Linear Fit @ {wavelengths[idx]:.0f} nm (R²={r_value**2:.2f}), τ₁/₂={-intercept/2*1/slope/60/60:.0f}h", color=colors[i][1], linewidth=2)
# Set axis labels and title
ax.set_title("Absorbance over Time at Selected Wavelengths", fontsize=16)
ax.set_xlabel("Time (s)", fontsize=14)
ax.set_ylabel("Absorbance", fontsize=14)
# Add legend and grid
ax.legend(fontsize=12)
ax.minorticks_on()
ax.grid(which='major', linestyle='-', linewidth=0.6, color='black', alpha=0.4)
ax.grid(which='minor', linestyle=':', linewidth=0.4, color='black', alpha=0.2)
# Final layout
ax.set_axisbelow(True)
plt.tight_layout()
plt.show()