Obtaining QY values for DAE#
and equivalent power for a given QY
Import the experiment Data#
import pandas as pd
exp_data = pd.read_csv("DATA/DAE.csv")
exp_data
| timestamp | cycle | type | 186.85486 | 187.31995223015844 | 187.78500323297956 | 188.250012996982 | 188.71498151068454 | 189.17990876260572 | 189.64479474126426 | ... | 1032.9632529736894 | 1033.3204920667242 | 1033.6776665334785 | 1034.034776362471 | 1034.3918215422202 | 1034.7488020612445 | 1035.1057179080635 | 1035.4625690711953 | 1035.8193555391586 | 1036.176077300472 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 2025-04-04 11:29:32.797890 | 1 | zero | 16.990556 | 24978.543889 | 249.154361 | 241.872694 | 253.523361 | 243.935833 | 258.499167 | ... | 652.315972 | 654.500472 | 655.471361 | 647.218806 | 664.694806 | 659.354917 | 666.151139 | 657.049056 | 657.049056 | 657.049056 |
| 1 | 2025-04-04 11:31:06.034842 | 1 | on | 16.990556 | 24978.543889 | 254.858333 | 231.395185 | 276.163951 | 261.600617 | 252.431111 | ... | 639.168519 | 672.070864 | 642.944198 | 664.789198 | 651.034938 | 650.495556 | 633.235309 | 639.707901 | 639.707901 | 639.707901 |
| 2 | 2025-04-04 11:31:07.075687 | 1 | on | 16.990556 | 24978.543889 | 240.025309 | 238.676852 | 256.206790 | 239.755617 | 265.106605 | ... | 653.192469 | 652.922778 | 643.483580 | 663.710432 | 668.834568 | 634.853457 | 628.111173 | 670.452716 | 670.452716 | 670.452716 |
| 3 | 2025-04-04 11:31:08.109308 | 1 | on | 16.990556 | 24978.543889 | 242.991914 | 246.767593 | 242.182840 | 280.479012 | 280.748704 | ... | 637.820062 | 659.665062 | 663.440741 | 645.641111 | 635.662531 | 629.189938 | 638.359444 | 631.347469 | 631.347469 | 631.347469 |
| 4 | 2025-04-04 11:31:09.153691 | 1 | on | 16.990556 | 24978.543889 | 241.643457 | 222.765062 | 271.039815 | 262.140000 | 255.667407 | ... | 652.113704 | 651.844012 | 651.574321 | 651.034938 | 648.877407 | 647.528951 | 638.629136 | 645.101728 | 645.101728 | 645.101728 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 614 | 2025-04-04 11:41:43.804611 | 1 | on | 16.990556 | 24978.543889 | 241.913148 | 236.789012 | 220.337840 | 231.934568 | 246.767593 | ... | 649.686481 | 641.326049 | 641.865432 | 641.865432 | 641.056358 | 645.371420 | 639.438210 | 631.347469 | 631.347469 | 631.347469 |
| 615 | 2025-04-04 11:41:44.858569 | 1 | on | 16.990556 | 24978.543889 | 212.786481 | 231.125494 | 234.361790 | 239.755617 | 265.106605 | ... | 638.089753 | 626.762716 | 630.268704 | 636.201914 | 634.853457 | 648.068333 | 654.001543 | 662.361975 | 662.361975 | 662.361975 |
| 616 | 2025-04-04 11:41:45.888795 | 1 | on | 16.990556 | 24978.543889 | 223.304444 | 240.564691 | 237.058704 | 251.622037 | 240.295000 | ... | 659.125679 | 635.392840 | 641.056358 | 646.719877 | 642.135123 | 654.540926 | 630.268704 | 648.338025 | 648.338025 | 648.338025 |
| 617 | 2025-04-04 11:41:46.925455 | 1 | on | 16.990556 | 24978.543889 | 232.204259 | 234.631481 | 235.170864 | 257.824938 | 243.800988 | ... | 621.368889 | 644.292654 | 632.156543 | 629.459630 | 631.617160 | 652.653086 | 630.808086 | 632.426235 | 632.426235 | 632.426235 |
| 618 | 2025-04-04 11:41:59.961864 | 1 | static | 16.990556 | 24978.543889 | 225.488944 | 217.964556 | 231.557000 | 247.455306 | 278.281028 | ... | 634.475889 | 630.228250 | 649.888750 | 648.675139 | 633.990444 | 646.247917 | 634.111806 | 643.213889 | 643.213889 | 643.213889 |
619 rows × 2051 columns
Get important data from dataframe#
import numpy as np
intensities=np.array(exp_data[(exp_data["type"] != "zero") & (exp_data["type"] != "static")].iloc[:, 3:], dtype=np.float64)
static=np.array(exp_data[(exp_data["type"]== "static")].iloc[:, 3:], dtype=np.float64)[0]
zero=np.array(exp_data[(exp_data["type"]== "zero")].iloc[:, 3:], dtype=np.float64)[0]
wavelengths = np.array(exp_data.columns[3:], dtype=np.float64)
timestamps = pd.to_datetime(exp_data["timestamp"][(exp_data["type"] != "zero") & (exp_data["type"] != "static")]) # Convert timestamp strings to datetime objects
timestamps = np.array((timestamps - timestamps.iloc[0]).dt.total_seconds()) # Convert to seconds since the first timestamp
def compute_absorbance(intensities: np.ndarray, static: np.ndarray, zero: np.ndarray) -> np.ndarray:
EPS = 1e-12
num = intensities - static
den = np.maximum(zero - static, EPS) # Éviter division par zéro
absorbance = -np.log10(np.maximum(num / den, EPS)) # Éviter log(0) ou log(négatif)
return absorbance
absorbance = compute_absorbance(intensities, static, zero)
Plotting#
Show code cell source
import matplotlib.pyplot as plt
import matplotlib.cm as cm
# Set global style
plt.style.use("ggplot") # Clean style
fig, axs = plt.subplots(1, 3, figsize=(18, 5))
fig.suptitle("Spectral Analysis", fontsize=16)
# --- Intensity Plot ---
axs[0].plot(wavelengths, intensities[0, :], label=f"I at t = {timestamps[0]:.0f} s", linewidth=2)
axs[0].plot(wavelengths, intensities[len(intensities)//2, :], label=f"I at t = {timestamps[len(intensities)//2]:.0f} s", linewidth=2)
axs[0].plot(wavelengths, intensities[-1, :], label=f"I at t = {timestamps[-1]:.0f} s", linewidth=2)
axs[0].plot(wavelengths, static, '--', label="Static", linewidth=2)
axs[0].plot(wavelengths, zero, '--', label="Zero", linewidth=2)
axs[0].set_title("Intensity over Wavelength")
axs[0].set_xlabel("Wavelength (nm)")
axs[0].set_ylabel("Intensity")
axs[0].legend()
axs[0].grid(True, which='major', linestyle='--', linewidth=0.6, color='gray', alpha=0.5, zorder=0)
axs[0].minorticks_on()
axs[0].grid(True, which='minor', linestyle=':', linewidth=0.3, color='lightgray', alpha=0.4, zorder=0)
# --- Absorbance Plot ---
axs[1].plot(wavelengths, absorbance[0, :], label=f"t = {timestamps[0]:.0f} s", linewidth=2)
axs[1].plot(wavelengths, absorbance[len(absorbance)//2, :], label=f"t = {timestamps[len(absorbance)//2]:.0f} s", linewidth=2)
axs[1].plot(wavelengths, absorbance[-1, :], label=f"t = {timestamps[-1]:.0f} s", linewidth=2)
axs[1].set_title("Absorbance over Wavelength")
axs[1].set_xlabel("Wavelength (nm)")
axs[1].set_ylabel("Absorbance")
axs[1].set_ylim(-0.4, 2)
axs[1].legend()
axs[1].grid(True, which='major', linestyle='--', linewidth=0.6, color='gray', alpha=0.5, zorder=0)
axs[1].minorticks_on()
axs[1].grid(True, which='minor', linestyle=':', linewidth=0.3, color='lightgray', alpha=0.4, zorder=0)
# --- Absorbance vs Time Plot ---
WL = [505, 562]
idxs = [np.argmin(np.abs(wavelengths - wl)) for wl in WL]
for i, idx in enumerate(idxs):
axs[2].plot(timestamps, absorbance[:, idx], label=f"{wavelengths[idx]:.0f} nm", linewidth=2)
axs[2].set_title("Absorbance over Time")
axs[2].set_xlabel("Time (s)")
axs[2].set_ylabel("Absorbance")
axs[2].legend()
axs[2].grid(True, which='major', linestyle='--', linewidth=0.6, color='gray', alpha=0.5, zorder=0)
axs[2].minorticks_on()
axs[2].grid(True, which='minor', linestyle=':', linewidth=0.3, color='lightgray', alpha=0.4, zorder=0)
plt.tight_layout(rect=[0, 0, 1, 0.95])
plt.show()
Clean Data :#
# Define wavelength range of interest
wavelength_range = [560, 565]
idx_range = np.argmin(np.abs(wavelengths[:, None] - wavelength_range), axis=0)
# Extract and normalize absorbance data within the wavelength range
absorbance_subset = absorbance[:, idx_range[0]:idx_range[1]]
absorbance_subset -= np.min(absorbance_subset)
wavelengths_subset = wavelengths[idx_range[0]:idx_range[1]]
# Filter out specific time points (e.g., first frame and last 300 entries)
# Adjust indices as needed
time_filter_indices = [0] + list(range(-300, 0))
absorbance_subset = np.delete(absorbance_subset, time_filter_indices, axis=0)
timestamps_filtered = np.delete(timestamps, time_filter_indices)
# Create subplots
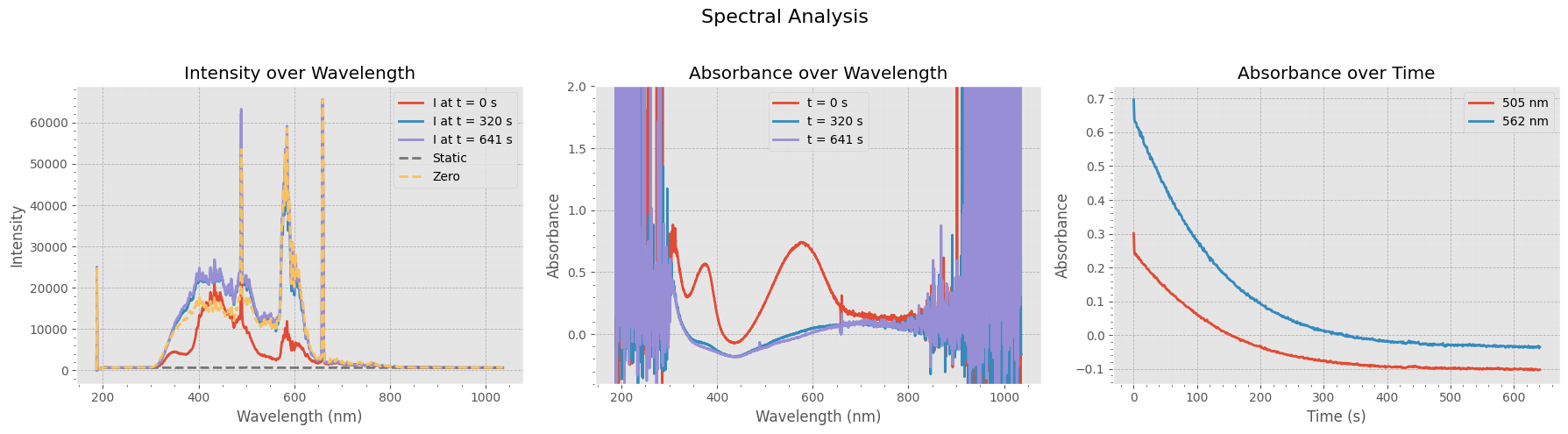
fig, axs = plt.subplots(1, 3, figsize=(18, 5))
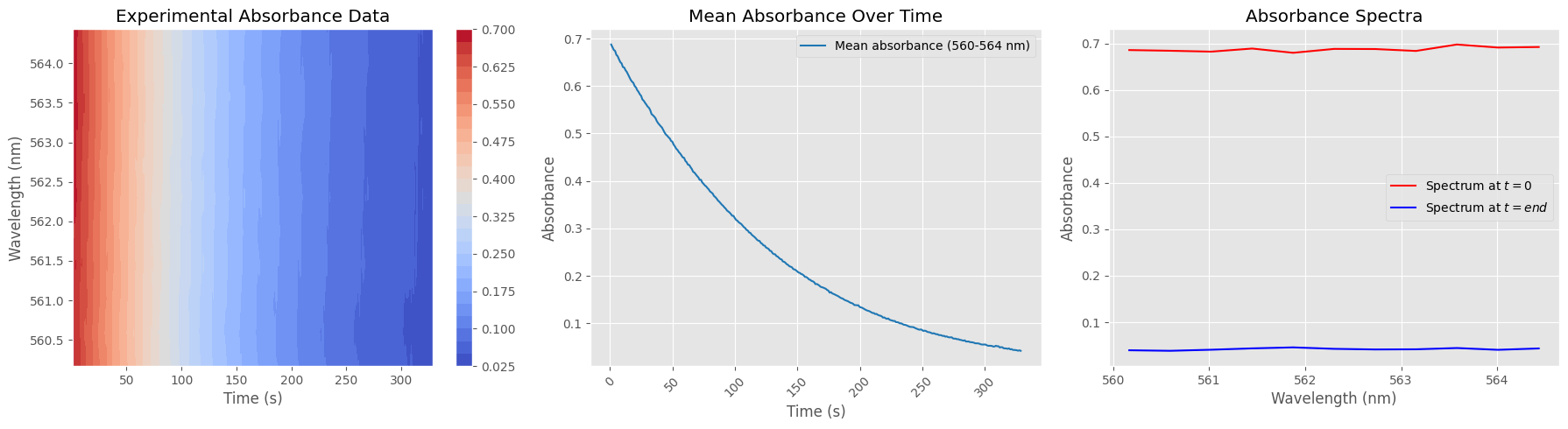
# --- 1. Absorbance Heatmap ---
X, Y = np.meshgrid(timestamps_filtered, wavelengths_subset)
contour = axs[0].contourf(X, Y, absorbance_subset.T, levels=30, cmap="coolwarm")
axs[0].set_title("Experimental Absorbance Data")
axs[0].set_xlabel("Time (s)")
axs[0].set_ylabel("Wavelength (nm)")
fig.colorbar(contour, ax=axs[0])
# --- 2. Absorbance Over Time (Averaged Across Wavelength Range) ---
mean_absorbance = np.mean(absorbance_subset, axis=1)
axs[1].plot(timestamps_filtered, mean_absorbance, label=f"Mean absorbance ({wavelengths_subset[0]:.0f}-{wavelengths_subset[-1]:.0f} nm)", color='tab:blue')
axs[1].set_title("Mean Absorbance Over Time")
axs[1].set_xlabel("Time (s)")
axs[1].set_ylabel("Absorbance")
axs[1].tick_params(axis="x", rotation=45)
axs[1].legend()
axs[1].grid(True)
# --- 3. Absorbance Spectrum at Start and End ---
axs[2].plot(wavelengths_subset, absorbance_subset[0, :], label="Spectrum at $t=0$", color='red')
axs[2].plot(wavelengths_subset, absorbance_subset[-1, :], label="Spectrum at $t=end$", color='blue')
axs[2].set_title("Absorbance Spectra")
axs[2].set_xlabel("Wavelength (nm)")
axs[2].set_ylabel("Absorbance")
axs[2].legend()
axs[2].grid(True)
plt.tight_layout()
plt.show()
Fit to get QY#
from scipy.integrate import solve_ivp
from scipy.optimize import curve_fit
# Physics constants :
h = 6.62607004 * 10 ** (-34)
NA = 6.02214086 * 10 ** (23)
c_vaccum = 299792458
wl = 505 # nm
v = c_vaccum / (wl * 1e-9)
volume = 14e-4 # L
l = 1 # cm
eps_closed_505 = 0.6e4 # M-1 . cm-1
eps_closed_562 = 1.1e4 # M-1 . cm-1
I_w_list = [14.21e-3*0.92, 14.27e-3]
for I_w in I_w_list:
C_exp = mean_absorbance/(eps_closed_562*l)
C0 = C_exp[0]
I_0 = I_w / (h * v * NA) / volume # mole de photons.L-1
print(I_0)
def f(t, C_closed, QY_CF2OF):
# Con[Con<0]=0
Iabs_CF = I_0 * (1 - np.exp(- eps_closed_505 * C_closed * l * np.log(10)))
dCCF_deri = -QY_CF2OF * Iabs_CF
# print(f"Con = {Con}, dC = {dCon_deri}, t = {t}")
return dCCF_deri
def fi(t, q1, c):
sol = solve_ivp(f, t_span, y0, args=(q1,), t_eval=timestamps_filtered)
if not sol.success:
print("⚠️ solve_ivp failed:", sol.message)
return None # Retourne None en cas d'échec
return sol.y.flatten()#+c
QY = 0.0233
initial_guess = (QY, 1)
bounds = ([0, -1], [1, 1])
t_span = [timestamps_filtered[0], timestamps_filtered[-1]]
y0 = np.array([C0])
# y = solve_ivp(f, t_span, y0, args=(QY,), t_eval=timestamps)
print("Reg start")
popt, pcov = curve_fit(fi, timestamps_filtered, C_exp, p0=initial_guess, bounds=bounds)
uncertainties = np.sqrt(np.diag(pcov))
# Print results
print("=== Fit Results ===")
for i, (coef, incert) in enumerate(zip(popt, uncertainties), start=1):
print(f"Parameter {i}: {coef:.4f} ± {incert:.4f}")
# Compute fitted values
fitted_values = fi(timestamps_filtered, *popt)
litt_values = fi(timestamps_filtered, QY,0)
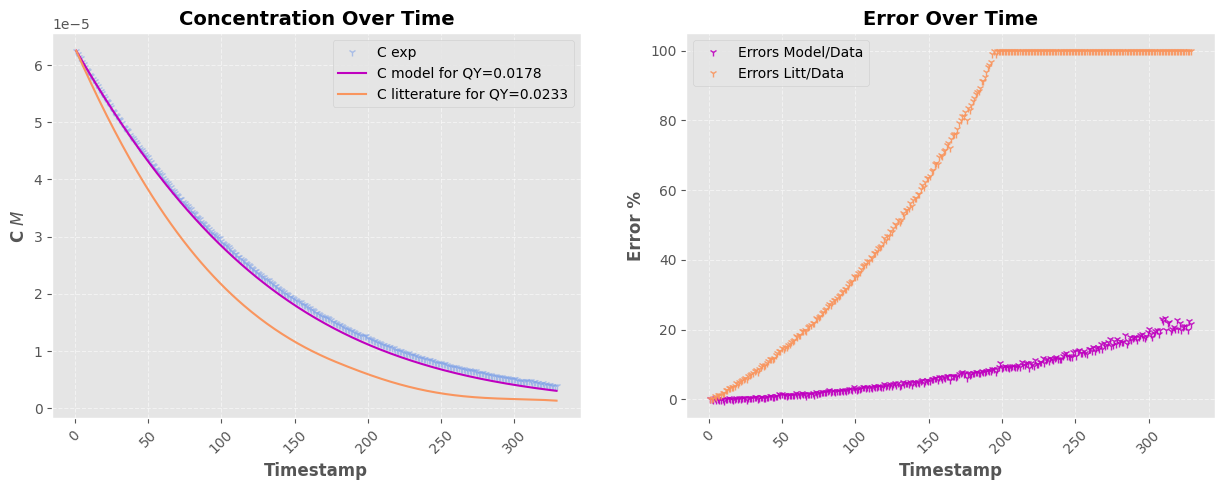
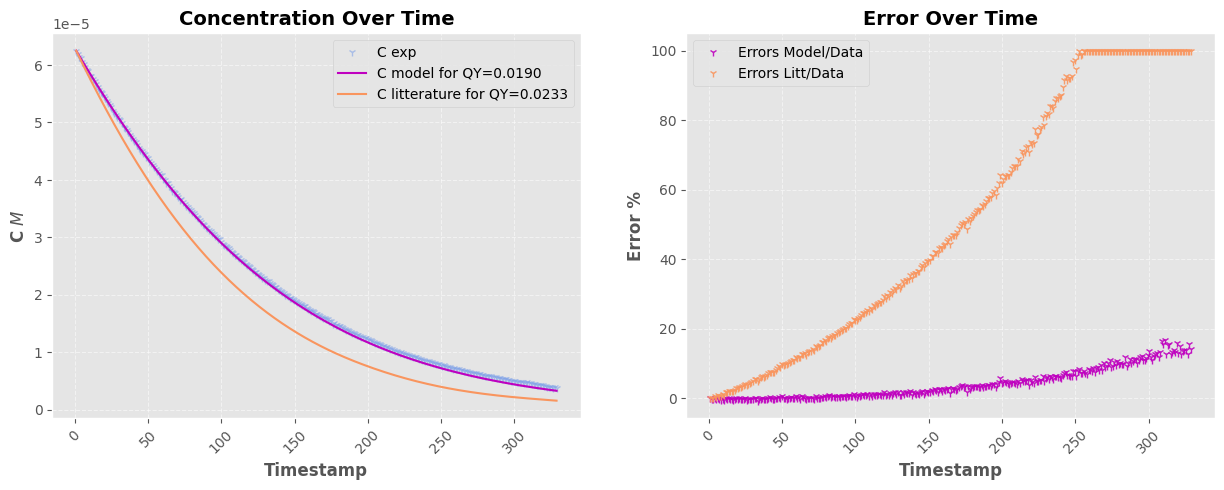
fig, a = plt.subplots(1, 2, figsize=(15, 5))
pastel_colors = ["m", "#f9955e", "#81a3e8"] # Mauve, orange, sky blue
# First Plot - Concentration Over Time
a[0].plot(timestamps_filtered, C_exp, "1", color=pastel_colors[2], label="C exp", alpha=0.6)
a[0].plot(timestamps_filtered, fitted_values, "-", color=pastel_colors[0], label=f"C model for QY={popt[0]:0.4f}")
a[0].plot(timestamps_filtered, litt_values, "-", color=pastel_colors[1], label=f"C litterature for QY={QY}")
a[0].set_xlabel("Timestamp", fontsize=12, fontweight="bold")
a[0].set_ylabel("C $M$", fontsize=12, fontweight="bold")
a[0].set_title("Concentration Over Time", fontsize=14, fontweight="bold")
a[0].tick_params(axis="x", rotation=45)
a[0].grid(linestyle="--", alpha=0.5)
a[0].legend()
# Second Plot - Error Over Time
a[1].plot(timestamps_filtered, (C_exp - fitted_values) * 100 / C_exp, "1", color=pastel_colors[0], label="Errors Model/Data", alpha=0.9)
ERR = (C_exp - litt_values) * 100 / litt_values
ERR[ERR > 100] = 100
ERR[ERR < -100] = -100
a[1].plot(timestamps_filtered, ERR, "1", color=pastel_colors[1], label="Errors Litt/Data", alpha=0.9)
a[1].set_xlabel("Timestamp", fontsize=12, fontweight="bold")
a[1].set_ylabel("Error %", fontsize=12, fontweight="bold")
a[1].set_title("Error Over Time", fontsize=14, fontweight="bold")
a[1].tick_params(axis="x", rotation=45)
a[1].grid(linestyle="--", alpha=0.5)
a[1].legend()
plt.show()
3.9420090134680676e-05
Reg start
=== Fit Results ===
Parameter 1: 0.0190 ± 0.0000
Parameter 2: 1.0000 ± 0.0000
4.302884421732194e-05
Reg start
=== Fit Results ===
Parameter 1: 0.0178 ± 0.0000
Parameter 2: 1.0000 ± 0.0000